Ethyl carbamate hydrolase mutant and application thereof
A kind of urethane and hydrolase technology, applied in the biological field, can solve the problem of low applicability and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Example 1: Agrobacterium tumefaciens d 3 Heterologous expression of amidase
[0021] (1) Use primer sequence D 3 -F,D 3 -R (Table 1) amplifies the amidase gene of Agrobacterium tumefaciens d3 (entrusted to Qingke Biosynthesis, the gene sequence is GenBank: AF315580, the amino acid sequence is shown in SEQ ID No.1), with primers pET30-F, pET30 -R amplification to obtain a linearized vector. Then use the ClonExpress II one-step cloning kit (purchased from Novizyme, Cat. No. C112-02) to connect the gene fragment between the NdeI and HindIII sites of the vector. The constructed plasmid was named pET-30a(+) / AtUTNase. The gene on the constructed plasmid has a His-tag for easy purification.
[0022] Table 1 Primers
[0023]
[0024]
[0025] (2) Transform the plasmid pET-30a(+) / AtUTNase into Escherichia coli BL21(DE3), inoculate the Escherichia coli BL21(DE3) carrying pET-30a(+) / AtUTNase into a test tube containing kanamycin and cultivate overnight , transferred t...
Embodiment 2
[0027] Embodiment 2: the enzyme activity detection method of ethyl carbamate hydrolase
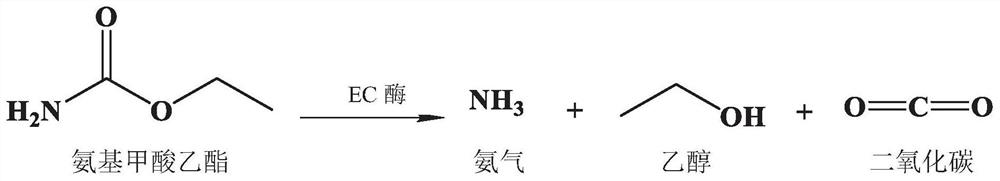
[0028] The principle of the method for detecting the enzyme activity of ethyl carbamate hydrolase is: under certain conditions, ethyl carbamate hydrolase decomposes ethyl carbamate to generate ethanol, ammonia and carbon dioxide. Ammonia reacts with phenol-sodium hypochlorite to form indophenol blue, which is blue, and its absorbance value is measured at 625nm with a spectrophotometer to analyze the amount of ammonia generated, so as to obtain the enzyme activity of ethyl carbamate hydrolase. Definition of enzyme activity: 1 μmol of ammonia produced by decomposing the substrate per minute under normal pressure and 37°C is an enzyme activity unit.
[0029] (1) Preparation of enzyme activity assay reagents:
[0030] Color developer I: Weigh 15g of phenol and 0.625g of sodium nitroferricyanide and dilute to 250mL with ultrapure water.
Embodiment 3
[0039] Embodiment 3: the purification of amidase
[0040] (1) Prepare the solution:
[0041] Binding Buffer: Weigh 1.7g of imidazole and 14.6g of NaCl in a beaker, add 50mL of 0.2M NaCl 2 HPO 4 -NaHPO 4 Buffer, use phosphoric acid to adjust the pH, and finally dilute to 500mL with deionized water.
[0042] Elution Buffer: weigh imidazole 5.1, 5.97, 6.8, 8.5, 9.35, 10.2, 17g and NaCl 14.6g in a beaker, add 50mL 0.2M Na 2 HPO 4 -NaHPO 4 For the buffer solution, use phosphoric acid to adjust the pH, and finally use deionized water to adjust the volume to 500mL, so as to prepare elution buffer solutions with imidazole concentrations of 150mM, 175mM, 200mM, 250mM, 275mM, 300mM, and 500mM, respectively.
[0043] (2) Operation process:
[0044] Pretreatment: 10 times column volume ddH 2 O washing nickel column, 10 times of column volume 50mM imidazole buffer equilibrium nickel column;
[0045] Adding samples: Repeat loading of the broken cell solution five times;
[0046] W...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com