Method for detecting diversity of large aquatic plants based on environmental DNA technology
A technology of aquatic plants and diversity, applied in the field of molecular ecology, can solve the problems of inaccurate estimation of abundance, easy neglect of species with secretive growth, identification errors, etc., and achieve the effects of accurate detection results, efficient detection, and simple sampling
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Embodiment 1, the establishment of the method for detecting the diversity of large aquatic plants based on environmental DNA technology
[0057] 1. Types of collected samples and screening of universal primers
[0058] 1. Universal primer design
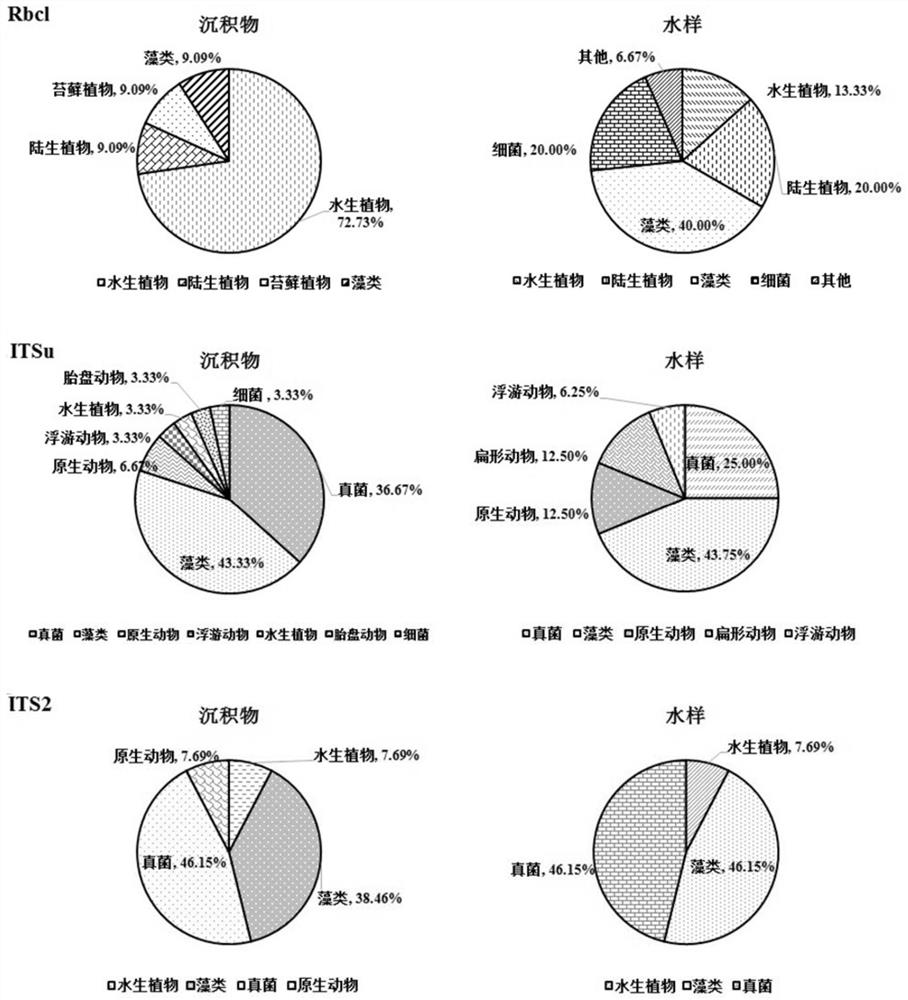
[0059] Choose chloroplast gene Rbcl to design Rbcl primer pair, this primer pair is made up of the upstream primer shown in sequence 1 and the downstream primer shown in sequence 2 (shown in table 1);
[0060] Choose chloroplast gene matK to design matK primer pair, this primer pair is made up of the upstream primer shown in sequence 3 and the downstream primer shown in sequence 4 (shown in table 1);
[0061] Ribosomal RNA (rRNA) gene ISu is selected to design ISu primer pair, and this primer is made up of the upstream primer shown in sequence 5 and the downstream primer shown in sequence 6 (shown in table 1);
[0062] Select ITS2 to design a primer pair for ITS2, which consists of an upstream primer shown in sequence 7 and ...
Embodiment 2
[0120] Embodiment 2, technical effect evaluation
[0121] In order to evaluate the reliability and effectiveness of the method in Example 1 (eDNA technology) in the survey of macrophyte diversity, the diversity of macrophyte in the Chaobai River Basin was investigated in combination with traditional survey methods. Based on the characteristics of species composition, relative abundance and community spatial distribution, the results of the two methods were compared.
[0122] 1. Sample collection
[0123] In June 2019, large-scale aquatic plant samples were collected in the Chaobai River Basin. A total of 9 sampling points were arranged, with 3 sampling points in the upper, middle and lower reaches, and 5 samples were collected in each sampling point ( Figure 4 , M1-M3; U1-U3; A1-A3).
[0124] eDNA method: sample collection is the same as the sample collection in Step 2 of Example 1.
[0125] Traditional method: through sample method combined with grass collector survey. S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com