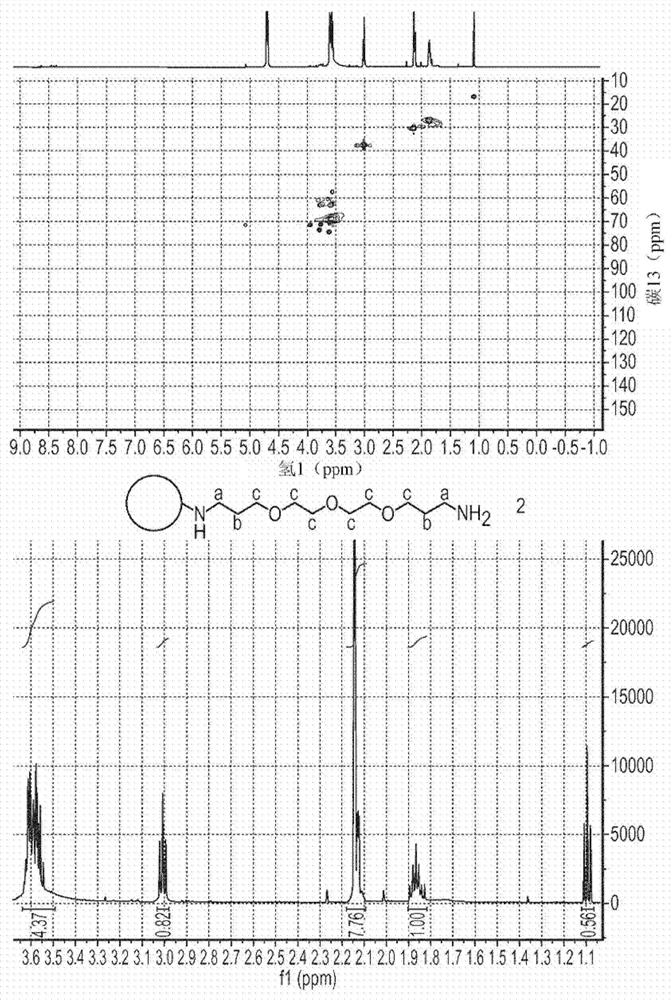
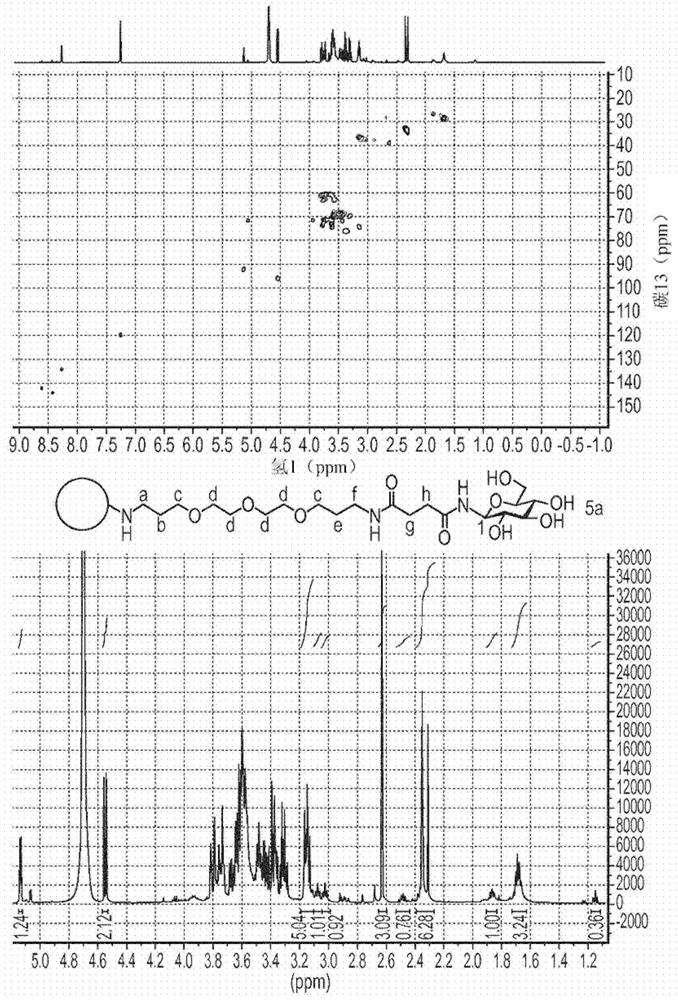
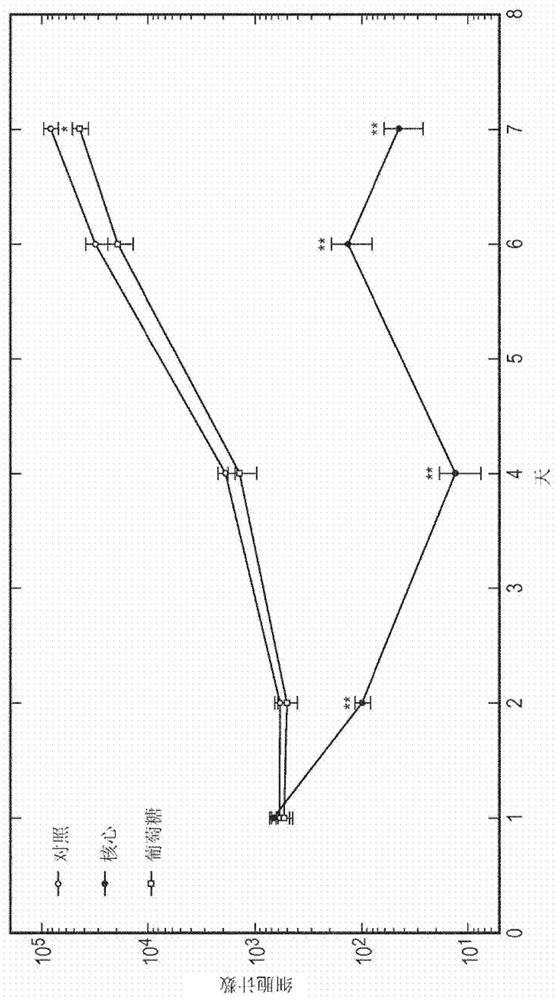
Compositions and methods for delivery of nucleic acid to plant cells
A plant and nucleic acid technology, applied in the field of nucleic acid delivery to plant cells, to achieve the effect of improving operational efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0182] Example 1 - Passivated carbon nanodot core formation
[0183] core synthesis
[0184] Combine glucosamine hydrochloride (1.00 g, 4.63 mmol) and 4,7,10-trioxa-1,13-tridecanediamine (TTDDA) (1.35 mL, 5.09 mmol) with 20 ml of ultrapure H 2 O mix. The mixture was then heated under microwave irradiation (3 minutes, 700 watts). 20mL CHCl 3 Added to the oil obtained and sonicated for 10 minutes. and discard the CHCl 3 . This process was repeated until the supernatant was clear.
[0185] The oil obtained after washing was then excluded using different conditions discussed below, depending on the scale of the reaction.
[0186] small scale
[0187] Then dissolve this brown oil in 20ml ddH 2 O and centrifuged through a 10,000 MWCO spin filter (GE Healthcare Life Sciences VIVASPIN 20, 5000 rpm, 1 hour).
[0188] The sample was then passed through a 200 nm syringe filter and ddH-treated with a Biotech cellulose ester membrane at 0.5 to 01 kDa MWCO 2 O dialyzed overni...
Embodiment 2
[0195] Example 2 - CND Core Functionalization and Nanocomplex Formation
[0196] CND-L-COOH
[0197] Succinic anhydride (5 g) was added to CND-L-NH 2 (1 g) (as obtained in Example 1) in distilled water (50 mL). The mixture was reacted overnight. The reaction volume was reduced under vacuum and CND-L-COOH was purified by dialysis against ultrapure water in 500 to 1000 MWCO overnight; or by eluting a size exclusion column (G15) with ultrapure water.
[0198] CND-L-PEG-NH 2
[0199] PEG1000-diamine (0.5 g) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (0.2 g) were added to CND-L-COOH (40 mg / mL) in distilled water ( 250 mL) and reacted overnight. The reaction volume was reduced under vacuum and CND-L-PEG1000-NH was purified by dialysis against ultrapure water in 1000 to 3000 MWCO overnight; or by eluting a size exclusion column (G15) with ultrapure water 2 . [PEG 1000 is a weight average molecular weight of 1000g.mol -1 of polyethylene glycol].
[0200] In separat...
Embodiment 3
[0207] Example 3 - Preparation of CND-plasmid polymer complex
[0208] hybrid approach
[0209] 10 μL of CND-L-PEG1000-NH as obtained in Example 2 2 (20 mg / mL), 10 μL of YFP229 plasmid, 130 μL of phosphate buffered saline (PBS) and 400 μL of ultrapure water, vortexed for 30 seconds and allowed to stand for 5 minutes before any analysis or experiment was performed.
[0210] In separate examples, the YFP229 plasmid was replaced with 131Y (7Kbp), PART27 (9Kbp), 127T (7Kbp) and 1T (7Kbp).
[0211] Precipitation
[0212] 10 μL CND-L-NH 2 (20 mg / mL) and 10 μL of Cas9 plasmid were vortexed for 30 seconds. The mixture was concentrated to dryness using a high speed vacuum concentrator and stored. The solids were resuspended in ultrapure water prior to any analysis or experiment.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com