Key protein identification method based on capsule neural network and ensemble learning
A neural network and ensemble learning technology, applied in the field of systems biology, can solve problems such as few features and no deep-level features, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
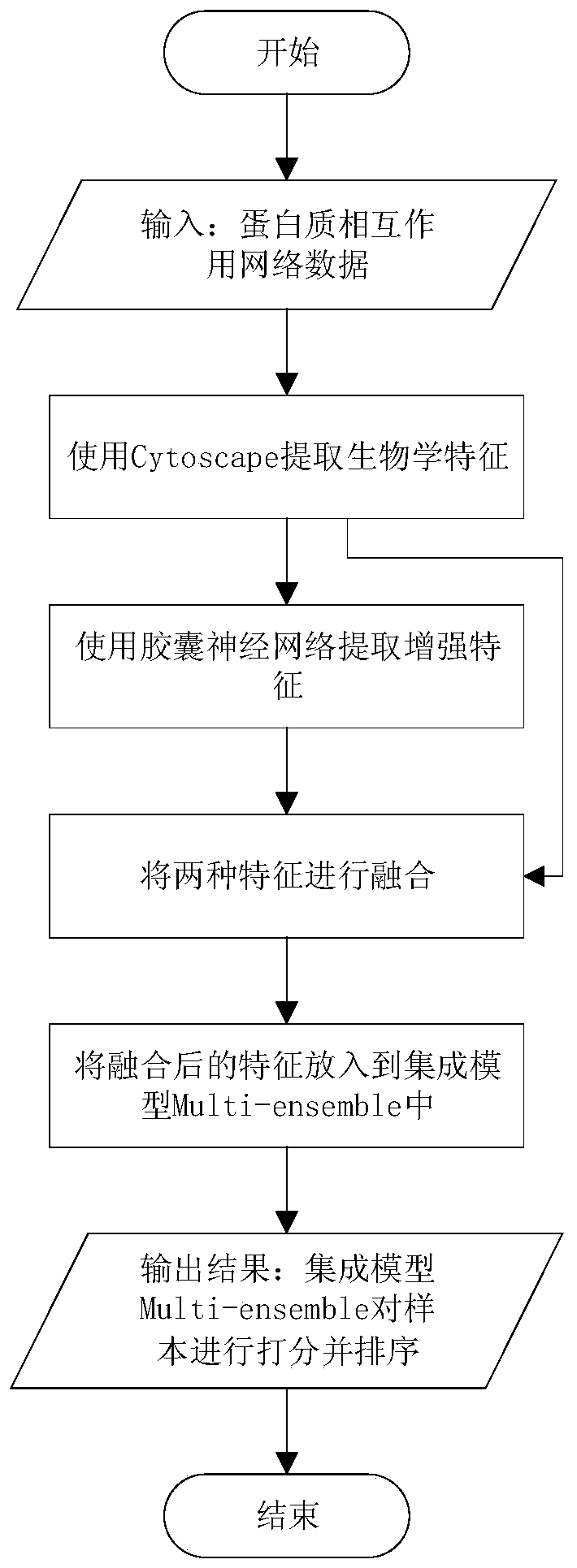
[0018] Embodiment 1: as Figure 1-2 As shown, a key protein identification method based on capsule neural network and integrated learning, the method steps are as follows:
[0019] Step 1: Use the Cytoscape tool to extract eight biological characteristics of proteins in the protein interaction network; among them, proteins are divided into two categories: non-key proteins and key proteins;
[0020] Key proteins tend to be the core nodes of PINs, because removing them would cause the PINs to collapse. The biological characteristics of proteins in the protein interaction network mainly include: betweenness centrality (Betweenness Centrality, BC), proximity centrality (Closeness Centrality, CC), degree centrality (Degree Centrality, DC), eigenvector centrality (Eigenvector Centrality, EC), Information Centrality (Information Centrality, IC), etc. The software Cytoscape was used to extract eight biological characteristics of the protein, BC, CC, DC, EC, IC, NC, SC and ION.
[0...
Embodiment 2
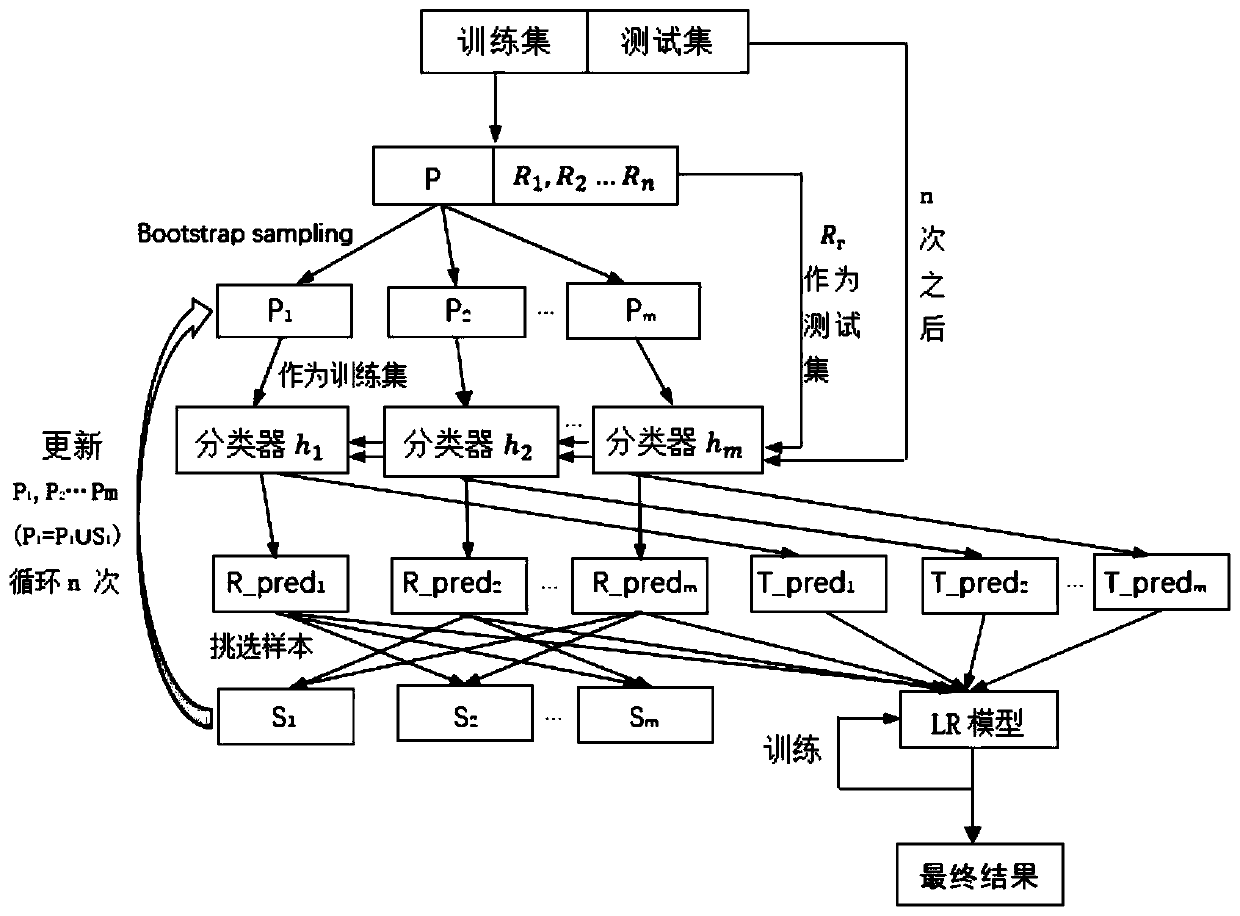
[0040] (1) To test the effectiveness of our invented method, we applied it to the Saccharomyces cerevisiae dataset and the E. coli dataset. Because they are well-studied and have the most complete and reliable key proteome and protein interaction network data across all species. The key proteins of Saccharomyces cerevisiae were obtained from the synthesis of MIPS database, SGD database, DEG database and SGDP database. Saccharomyces cerevisiae protein interaction network data were obtained from the DIP database. The protein interaction network of yeast consists of 5093 proteins and 24743 edges. Among the 5093 proteins in yeast, there were 1167 key proteins and 3926 non-key proteins, and the ratio of key proteins to non-key proteins reached 1:3.36. The key proteins of Escherichia coli were obtained from the DEG database. The protein interaction network data of Escherichia coli was downloaded from the DIP database, which included 2727 proteins and 11803 edges. Among the 2727 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com