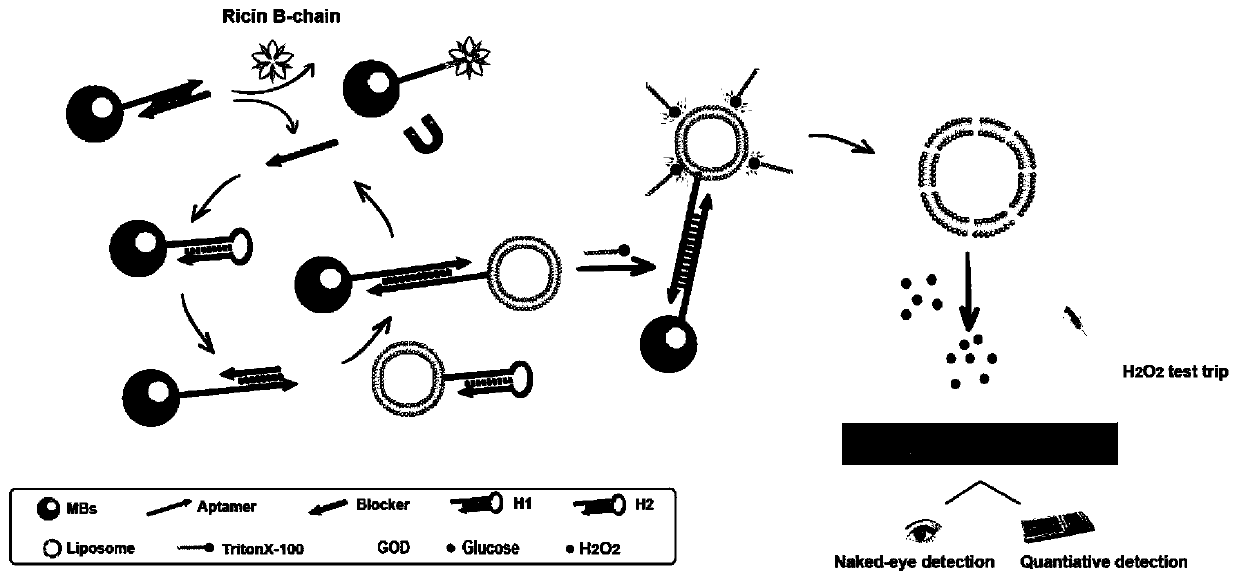
Method for detecting chain B of ricin in real time by using dual amplification strategy of catalytic hairpin assembly mediated lipidosome encoding magnetic beads
A ricin, double-amplification technology, applied in biochemical equipment and methods, material analysis by observing the impact on chemical indicators, microbial determination/inspection, etc., can solve the method of time-consuming, high immunogenicity, Complicated operation and other problems, to achieve the effect of convenient operation, high sensitivity and long time consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
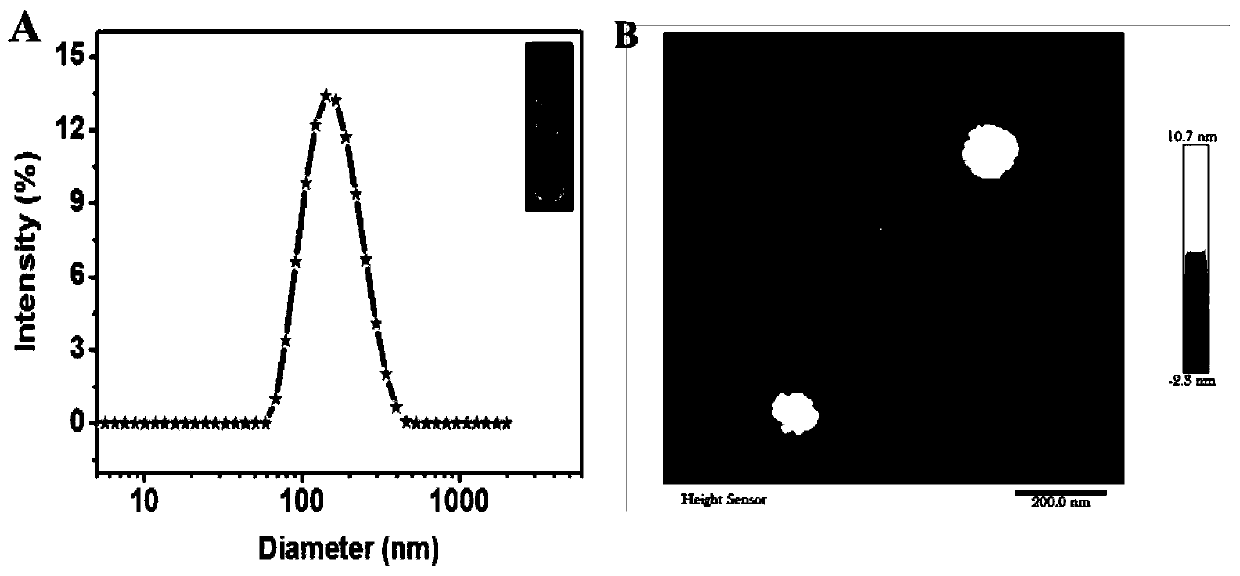
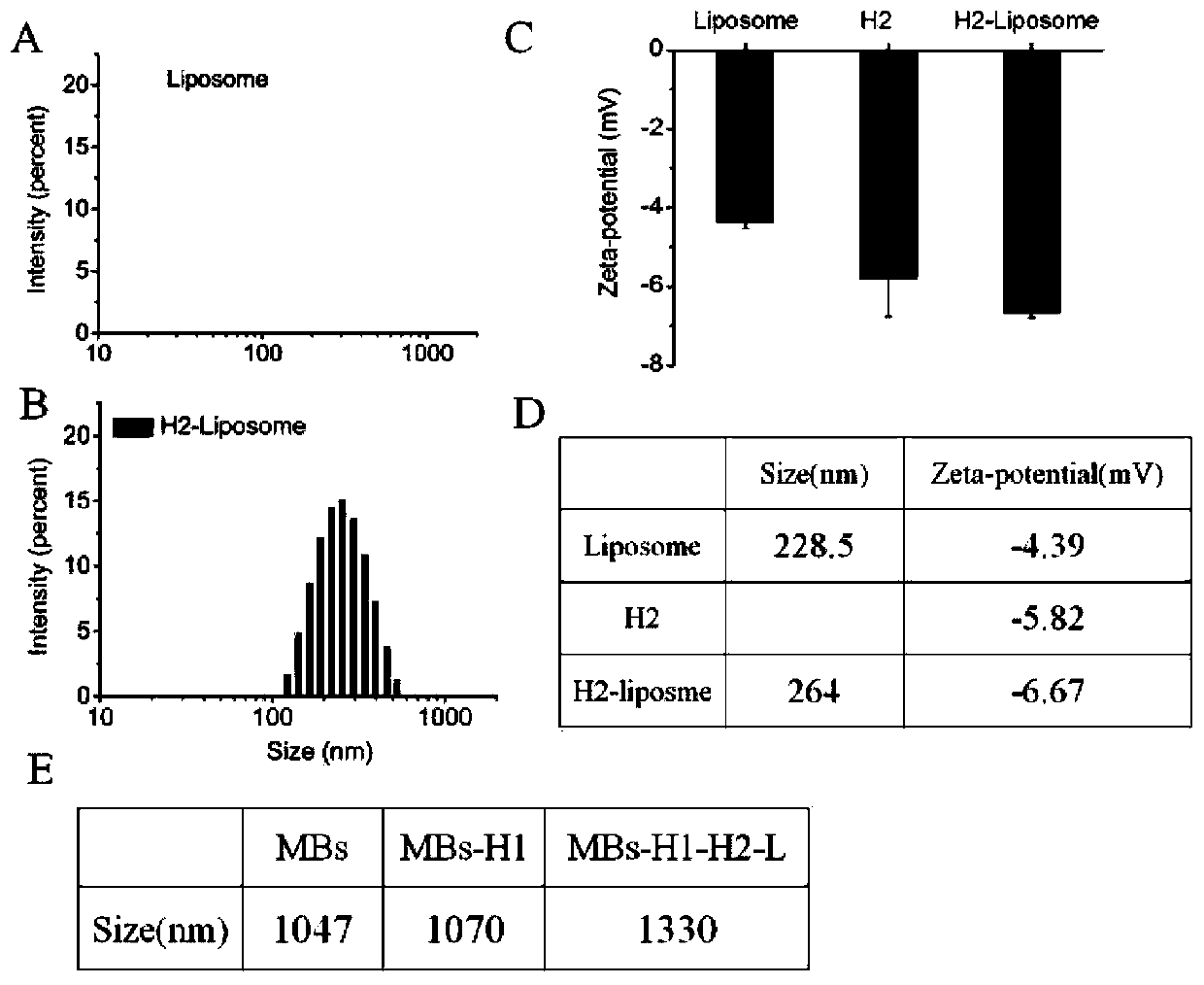
[0046] Example 1: Preparation and Characterization of Glucose Oxidase Liposomes
[0047] First, soybean lecithin, cholesterol and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (DSPE-PEG 2000 ) respectively according to 18mg, 4.6mg, 3.3mg (molar ratio 100:50:5) into the round bottom flask and dissolved with 1mL chloroform. Then, chloroform was removed under reduced pressure in a rotary evaporator at 40° C. for 30 minutes to obtain a film, and then vacuum-dried for 2 hours to remove residual organic solvent. Then, 2 mL of PBS (0.01 M, pH 7.4) solution containing 5 mg / mL GOD was added to the dried film, hydrated at 40 °C for 30 minutes to form a cloudy suspension, and then passed through a 100 nm polycarbonate membrane to obtain liposomes. Finally, a 300 kDa dialysis bag was used to stir in PBS (0.01 M, pH 7.4) at 4° C. for 24 h to remove uncoated glucose oxidase. Meanwhile, the obtained liposomes were stored at 4°C for subsequent use. T...
Embodiment 2
[0048] Embodiment 2: Preparation of magnetic beads-nucleic acid aptamer-blocker hybridization probe and MBs-H1 and H2-L GOD
[0049] (1) Preparation of magnetic bead-adapter-blocker hybridization probe: First, add the nucleic acid aptamer of ricin B chain, blocker and 10×TNaK solution together, and anneal the mixture at 95°C for 5 minutes Let stand for 30 minutes to cool to room temperature. Subsequently, streptavidin-modified magnetic beads (resuspended in 1×TNaK solution) were added to the above mixture and incubated at room temperature for 30 min. After removing the supernatant, the magnetic beads (MBs-aptamer-blocker ) three times and resuspended in 1×TNaK buffer.
[0050] (2) Preparation of MBs-H1: First, heat the biotin-modified H1 at the 3' end to 95°C for 1 minute and then let it stand for 30 minutes to cool to room temperature, then add streptavidin-modified magnetic beads (resuspended in 1×TNaK solution) and incubate at room temperature for 30 minutes with shaking...
Embodiment 3
[0052] Example 3: CHA-mediated liposome-encoded magnetic bead strategy for the detection of RTB
[0053] Add RTB and TNaK buffer to the magnetic bead-aptamer-blocker hybridization probe, incubate at room temperature for 40 minutes, and magnetically separate the supernatant, which is the blocker, and participate in the next step of the experiment. Then, MBs-H1, H2-L GOD MBs-H1-H2-L obtained after reacting with blocker at 37°C for 1 hour, washing with PBS three times to remove unreacted components GOD . Subsequently, 10 mM glucose solution (1% Triton X-100, 10 mM PBS, pH 5.5) was added to MBs-H1-H2-L GOD In , after reacting at 55 °C for 40 minutes, the liposome membrane was destroyed to release GOD, which catalyzed the oxidation of glucose to produce H 2 o 2 . Finally, through PTS and handheld H 2 o 2 The detector can realize naked-eye qualitative and quantitative detection of B chain at the same time.
[0054] See Table 1 for the nucleotide sequences involved in the pre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com