Genetic engineering bacterium highly producing D-allulose and application of genetic engineering bacterium
A technology of genetically engineered bacteria and psicose, applied in the field of bioengineering, can solve the problems of low yield, many by-products, difficult separation and purification, etc., and achieve the effect of reducing production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: DTEase gene target fragment amplification
[0030] By PCR method to DTEase-pMD TM The 18-T vector is used as a template, and DTE-F and DTE-R are used as primers to amplify and obtain the target fragment product of DTEase gene;
[0031] The primer sequences are as follows:
[0032] DTE-F: 5'-TGCACTGCAGATGTTGATATCCCCCTTTCGAG-3'(Pst I),
[0033] DTE-R: 5'-GGAAGCTTTCAGGCGTTCCGTGCGGC-3' (Hind III).
[0034] The PCR reaction system is as follows:
[0035]
[0036] The PCR reaction program was as follows: pre-denaturation at 95°C for 5 min; 35 cycles of 98°C for 10 s, 56°C for 15 s, and 72°C for 1 min; extension at 72°C for 5 min. Finally, the target fragment of the DTEase gene (as shown in SEQ ID NO.1) was obtained through amplification.
Embodiment 2
[0037] Example 2: Cloning vector DTEase- -Build of T1
[0038] The target fragment of the DTEase gene obtained by amplification and -T1 (Beijing Quanshijin Biotechnology Co., Ltd.) performs the ligation reaction according to the following system:
[0039]
[0040] The reaction process is as follows: Gently mix the reaction mixture, react in a PCR instrument at a temperature of 25°C for 10 minutes, and place in an ice bath for 2 to 3 minutes after the reaction.
[0041]Add the ligation product to 100 μL Trans1-T1 competent cells (add the ligation product when the competent cells are just thawed), flick and mix well, and keep in an ice bath for about 30 min; heat shock in a water bath at 42°C for 30 s, and immediately place on ice for 2 min; add 250 μL of SOC medium equilibrated to room temperature, and incubate at 200 rpm at 37°C for 1 h; take 200 μL of the bacterial solution and spread it on LB solid medium (containing 50 μg / mL Kan), and incubate overnight at 37°C. Pic...
Embodiment 3
[0045] Embodiment 3: Construction of expression vector DTEase-pP43NMK
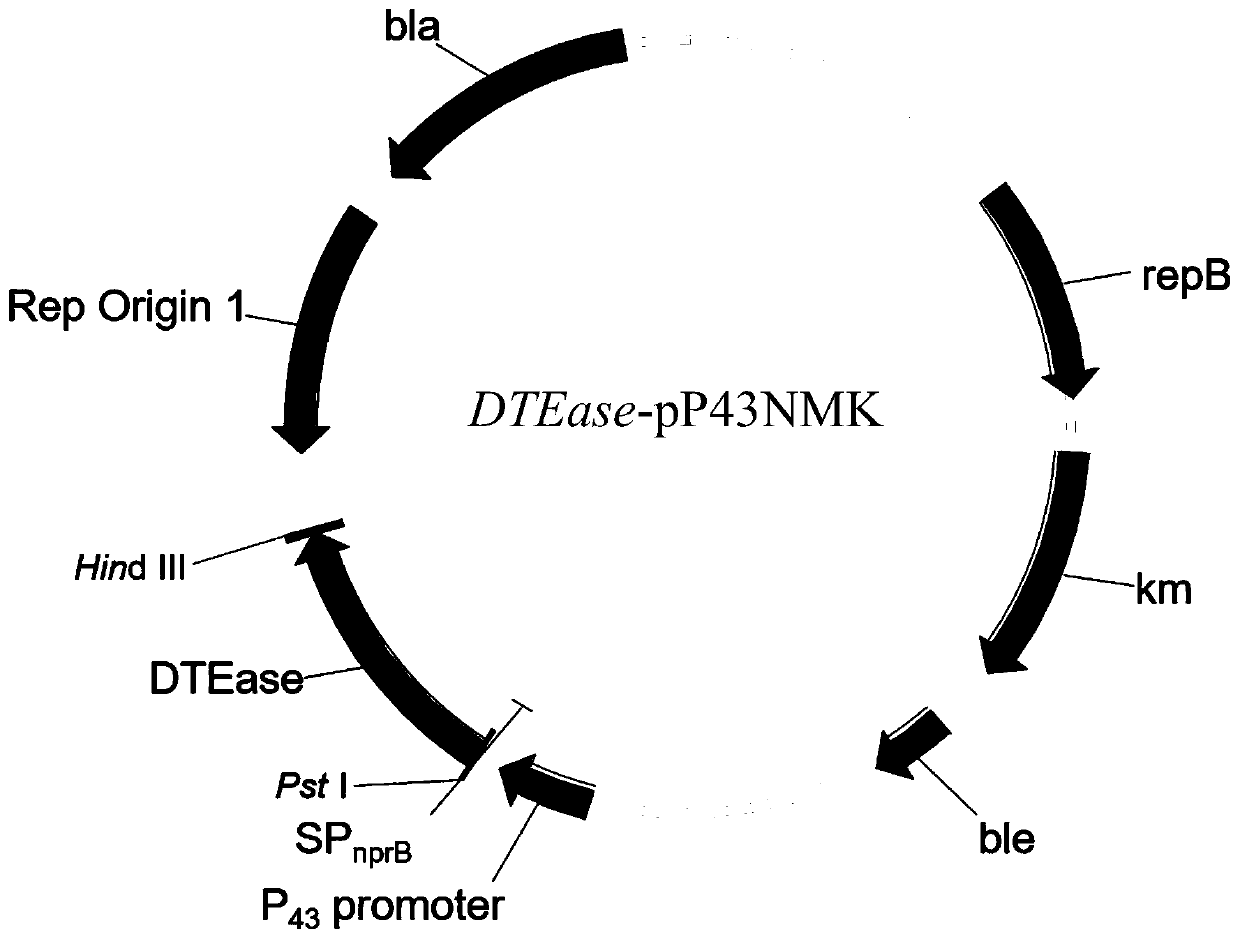
[0046] The plasmid DTEase- -T1 and pP43NMK were digested with Pst I and Hind III respectively, and the gene fragments were recovered from the gel, and then transformed into E.coli DH5α competent cell (the specific method is the same as in Example 2 -T1 transformation process), spread on LB plates containing Amp (50 μg / mL), and culture overnight at 37°C. Pick positive clones, use DTE-F and DTE-R as primers to screen positive clones by bacterial liquid PCR, extract plasmids, double enzyme digestion (Pst I and Hind III) and sequence verification. The recombinant plasmid verified and sequenced correctly was named as DTEase-pP43NMK ( figure 1 ).
[0047] The ligation reaction system of the linearized gene fragment kit is as follows (target fragment: linearized vector = 1:100 or more):
[0048]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com