Minimalycin Biosynthetic Gene Cluster, Recombinant Bacteria and Their Applications
A biosynthesis and gene cluster technology, applied in the field of microbial gene resources and genetic engineering, can solve the problem of little understanding of the biosynthetic pathway of minomycin, and achieve the effect of increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
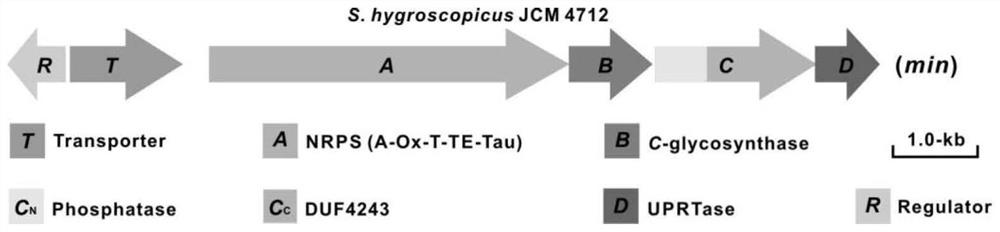
[0041] Example 1 Cloning and Analysis of the Biosynthetic Gene Cluster of Minimalycin
[0042] (1) First extract the total DNA of Streptomyces hygroscopicus JCM 4712: Take 30 μL of spores of Streptomyces hygroscopicus JCM 4712 into 50 mL of TSB medium, culture at 30°C, 200 rpm, for 24-36 hours, until the medium becomes turbid. Take 50mL of Streptomyces hygroscopicus JCM 4712 bacteria solution, centrifuge at 4000rpm, 4°C, 10min to remove the supernatant, and collect the bacteria. Dissolve the bacteria in 25mL of 10.3% sucrose solution, shake and mix to wash the bacteria, centrifuge at 4000rpm, 4°C, 10min to remove the supernatant; then dissolve the bacteria in 15mL of setbuffer, shake and mix, 4000rpm, 4°C, Centrifuge for 10 minutes to remove the supernatant, and repeat twice; dissolve the bacteria in 10 mL of setbuffer, shake and mix well, add 50 μL of lysozyme solution (100 mg / mL) and place it in a 37°C water bath for 30 minutes; then add 280 μL of proteinase K Solution (50m...
Embodiment 2
[0047] Example 2 In vivo functional determination of minimalmycin biosynthesis-related genes
[0048] (1) Construction of a recombinant plasmid containing the minimalmycin biosynthetic gene cluster in vitro: We infer that the gene cluster responsible for the minimalmycin biosynthesis is about 11.2kb, and through sequence analysis, it is found that the gene cluster responsible for the minimalmycin biosynthesis is in the middle It comes with a BglII enzyme cutting site, so the present invention divides the gene cluster into two parts (5290bp and 6089bp in size respectively) with BglII as the dividing point for PCR amplification, and the fragment with a size of 5290bp is added with SpeI at the 5' end. Restriction site, the 6089-bp fragment has an EcoRI restriction site added at the 3' end. Two fragments were respectively amplified using the total DNA of Streptomyces hygroscopicus JCM 4712 as a template. The 5290-bp fragment was digested with SpeI and BglII, and the 6089-bp fragme...
Embodiment 3
[0052] Example 3 In vitro functional verification of the non-ribosomal polypeptide synthase MinA encoded by the minamycin biosynthesis-related gene minA
[0053] The minA gene was amplified by PCR and then cloned into an expression vector, then transformed into E. coli BAP1, heterologously overexpressed and purified in E. coli, and analyzed by SDS-PAGE to obtain a relatively pure protein MinA. The specific operation Instructions: ①Pick positive single clones and culture overnight at 37°C in 5mL LB medium, transfer to 500mL LB at 1%, and culture at 37°C until the cell OD 600 To 0.5-0.8, add IPTG (final concentration 0.1-0.2mM) to induce, incubate at 18°C for 20h, centrifuge at 6000rmp for 15min to collect the cells; After the bacteria were mixed, the Escherichia coli cells were sonicated with an ultrasonic breaker, centrifuged at 12000 rpm at 4°C for 20 minutes to take the supernatant, and at 4°C, the supernatant was put into a gravity column with nickel filler, and used with...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com