Expression vector for observing localization of bacterial protein and construction method of expression vector
A technology for expressing vectors and bacterial proteins, which is applied in the biological field, can solve the problem of fewer prokaryotic protein positioning vectors, and achieve a strong inclusive effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1, the construction of expression vector
[0036] S1. Construction of expression vector pBBRlacITac-gfp and fluorescence detection
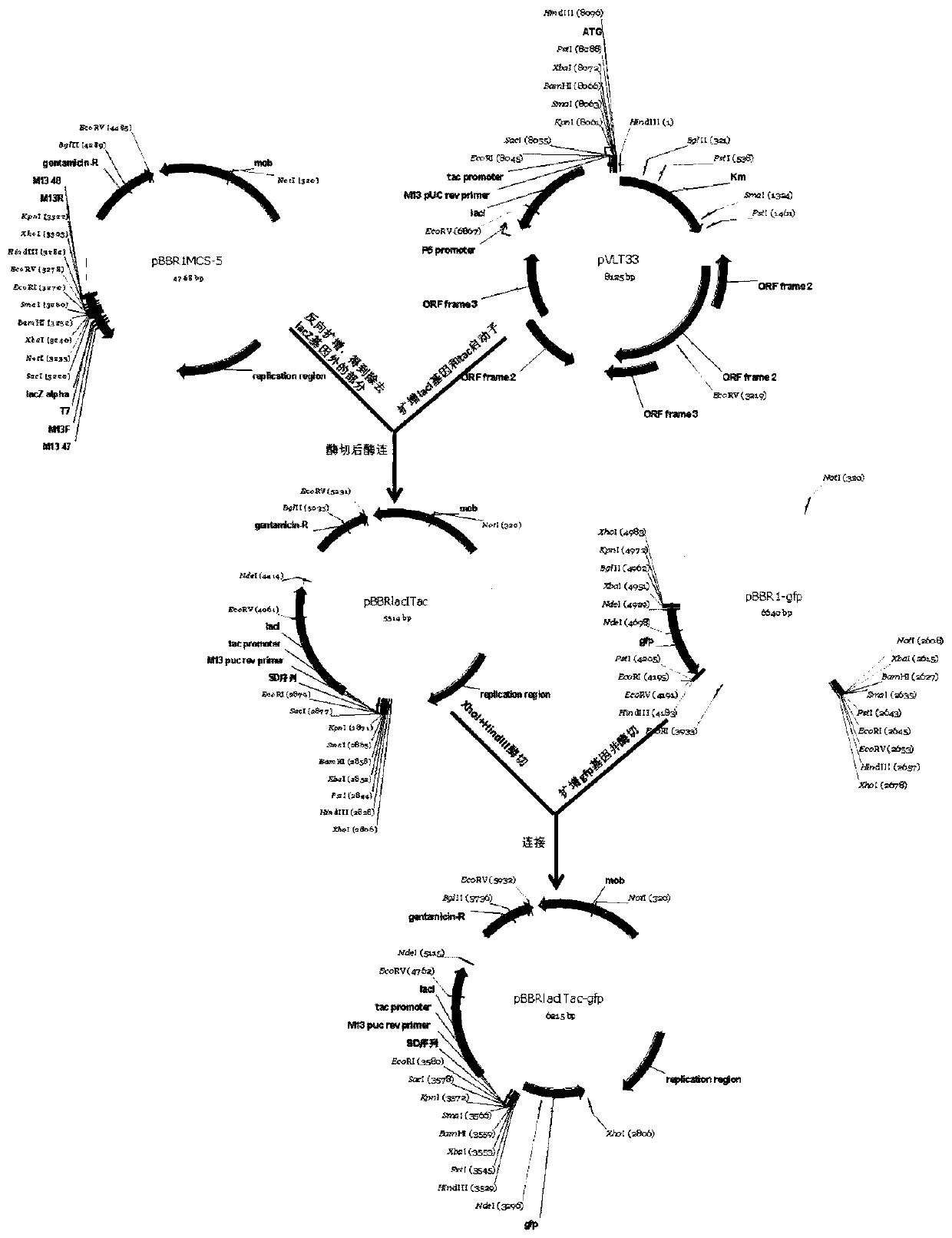
[0037] The schematic diagram of the construction of the expression vector pBBRlacITac-gfp is shown in figure 1 As shown, use the primer pair P1 / P2 and use the pBBR1MCS-5 plasmid as a template to reversely amplify the fragments on pBBR1MCS-5 except for lacZ; use the primer pair P3 / P4 to use the pVLT33 plasmid as a template to amplify the lacI gene and tac promoter; the above two PCR products were digested with NdeI and XhoI, ligated with T4 DNA ligase, and sequenced to obtain the vector pBBRlacITac. Use the primer pair P5 / P6 to amplify the gfp gene using the pBBR1-gfp plasmid as a template; digest the gfp gene with restriction enzymes XhoI+HindIII, and then use T4 DNA ligase to connect it to the pBBRlacITac vector that has been cut with the same enzyme , Sequencing verification, confirming that the gfp gene sequence is correct,...
Embodiment 2
[0041] Embodiment 2, utilize MotA protein as target protein to detect the feasibility of the present invention
[0042] S1. Construction of expression vector pBBRlacITac-motA-gfp
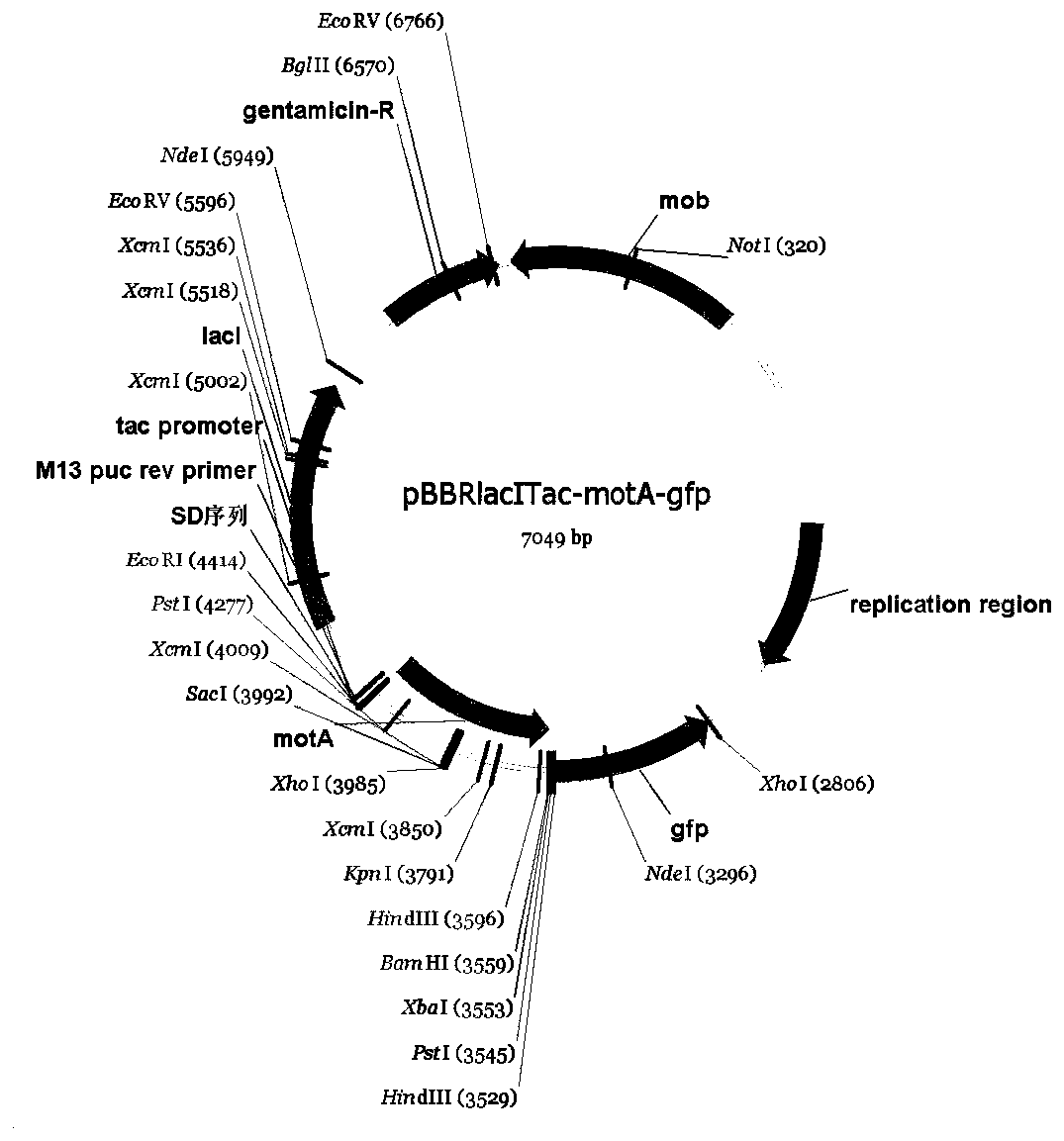
[0043] The map of the expression vector pBBRlacITac-motA-gfp is as follows image 3 shown. Pseudomonas putida KT2440 is a monotendex cluster flagellate. MotA is a KT2440 flagellar matrix component protein that localizes to bacterial endpoints. The present invention uses the pBBRlacITac-gfp carrier to detect the spatial location of MotA in KT2440 to verify whether the carrier has expected functions. Using the total genomic DNA of Pseudomonas putida KT2440 as a template, the motA gene was amplified using the primer pair P7 / P8, then ligated into the pBBRlacITac-gfp vector with EcoRI and BamHI, and sequenced to verify that the motA gene was inserted into the pBBRlacITac-gfp vector Finally, the expression vector pBBRlacITac-motA-gfp was obtained. The motA coding region amplified by PCR does not have...
Embodiment 3
[0046] Embodiment 3, utilize KatG protein as target protein to detect the feasibility of the present invention
[0047] S1. Construction of expression vector pBBRlacITac-katG-gfp
[0048] The map of the expression vector pBBRlacITac-katG-gfp is as follows Figure 5 shown. KatG is a kind of catalase in Escherichia coli, which can hydrolyze hydrogen peroxide in cells and relieve oxidative stress crisis. The expression of KatG was induced by hydrogen peroxide. If no hydrogen peroxide or other oxidative stress substances were added to the medium, KatG could only be expressed in a small amount in the late growth stage of E. coli, and it was difficult to observe its spatial location. The present invention observes the spatial localization of KatG in the logarithmic growth phase of Escherichia coli under the condition of not adding hydrogen peroxide. Using Escherichia coli K-12 genomic DNA as a template, the primer pair P9 / P10 was used to amplify the katG gene, and the XbaI and H...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com