Application and fixed-point knockout method of EMC3 gene
A targeted knockout and gene knockout technology, applied in applications, other methods of inserting foreign genetic materials, genetic engineering, etc., to achieve the effect of inhibiting JEV replication and improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
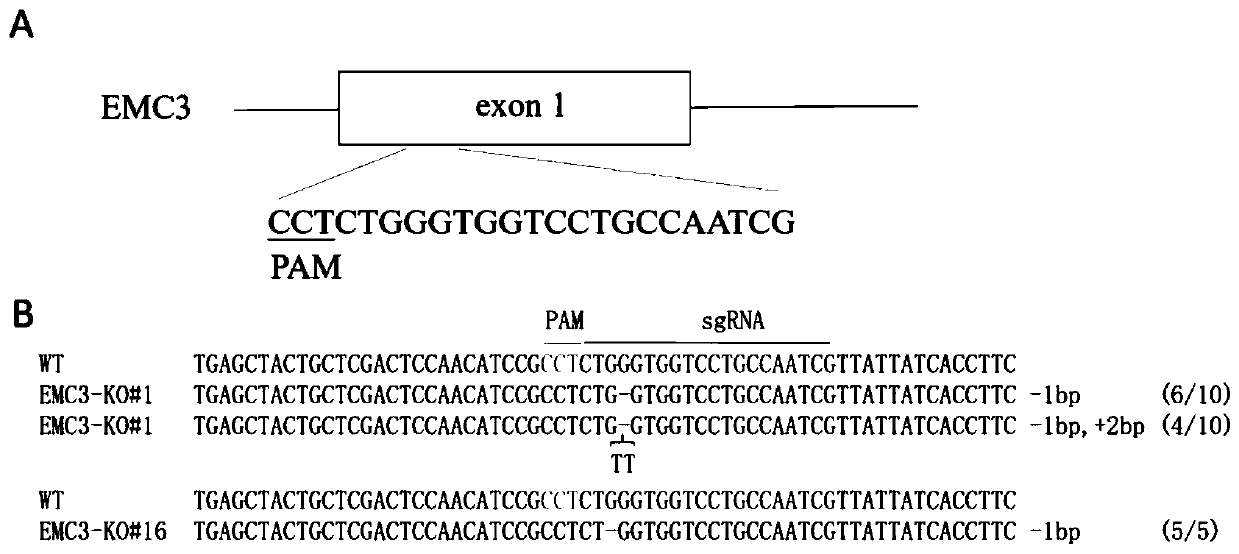
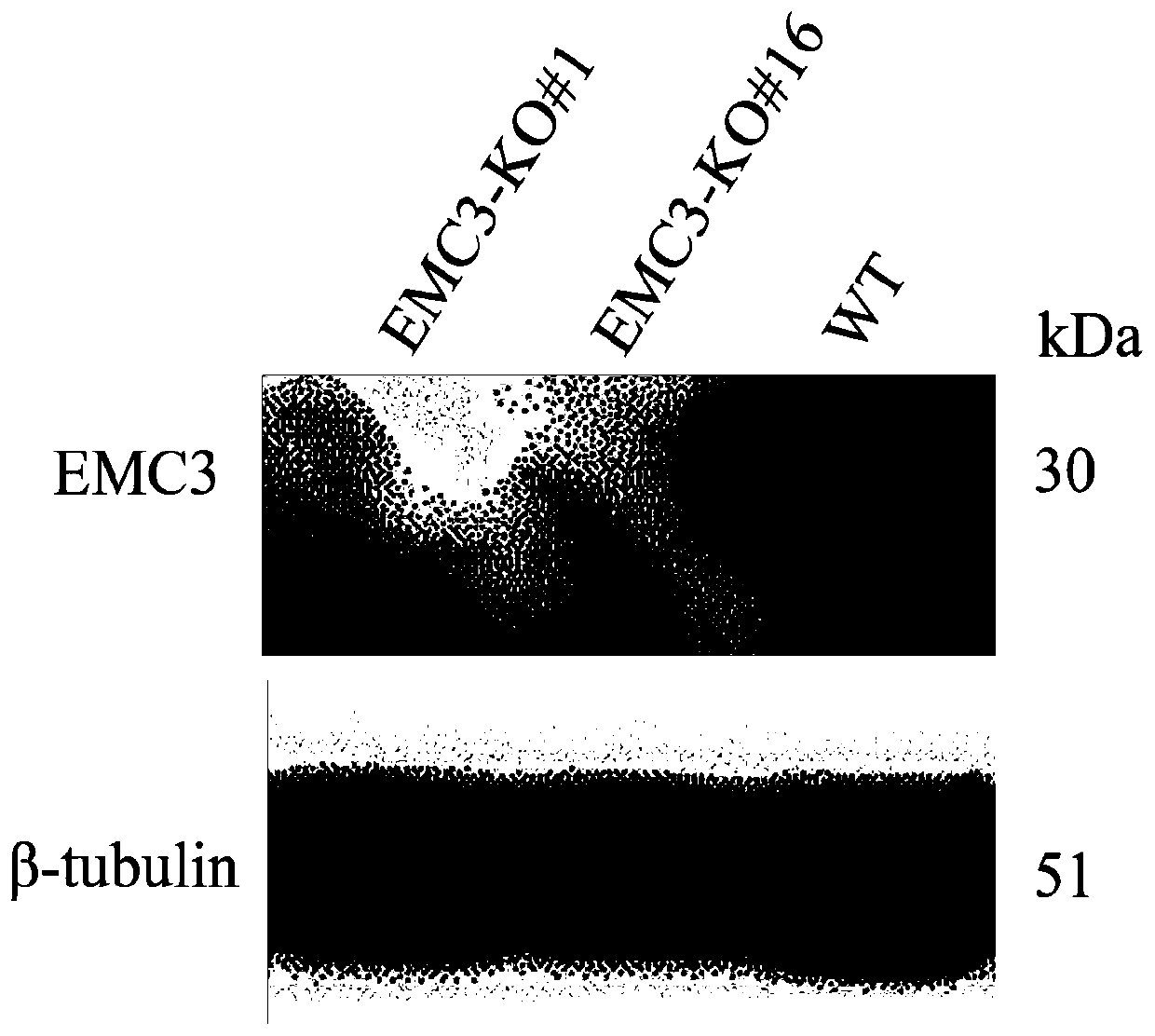
[0037] Example 1: Construction of EMC3 gene knockout cell lines using gene editing technology
[0038] First, download the porcine EMC3 gene exon sequence (accession number: ENSSSCG00000011562.3) and the whole pig genome sequence (version number: Sus_scrofa.Sscrofa11.1) from the ensemble database (www.ensembl.org), and then, use sgRNAcas9 The software (www.biootools.com) designed the sgRNA targeting the porcine EMC3 gene (Table 1), and selected the best sgRNA according to the specificity evaluation results. The sgRNA ID was EMC3_A_47, and the target sequence was "CGATTGGCAGGACCACCCAGAGG". It does not have 1 and 2 base mismatched off-target sites ( figure 1 A).
[0039] Table 1 Software design and evaluation of sgRNA targeting porcine EMC3 gene
[0040]
[0041]
[0042] Further, based on the lenti-sgRNA-EGFP lentiviral vector as the backbone, design and synthesize sgRNA primers, as follows EMC3-sgRNA-F: 5′-caccgCGATTGGCAGGACCACCCAG-3′, EMC3-sgRNA-R: 5′-aaacCTGGGTGGTCCT...
Embodiment 2
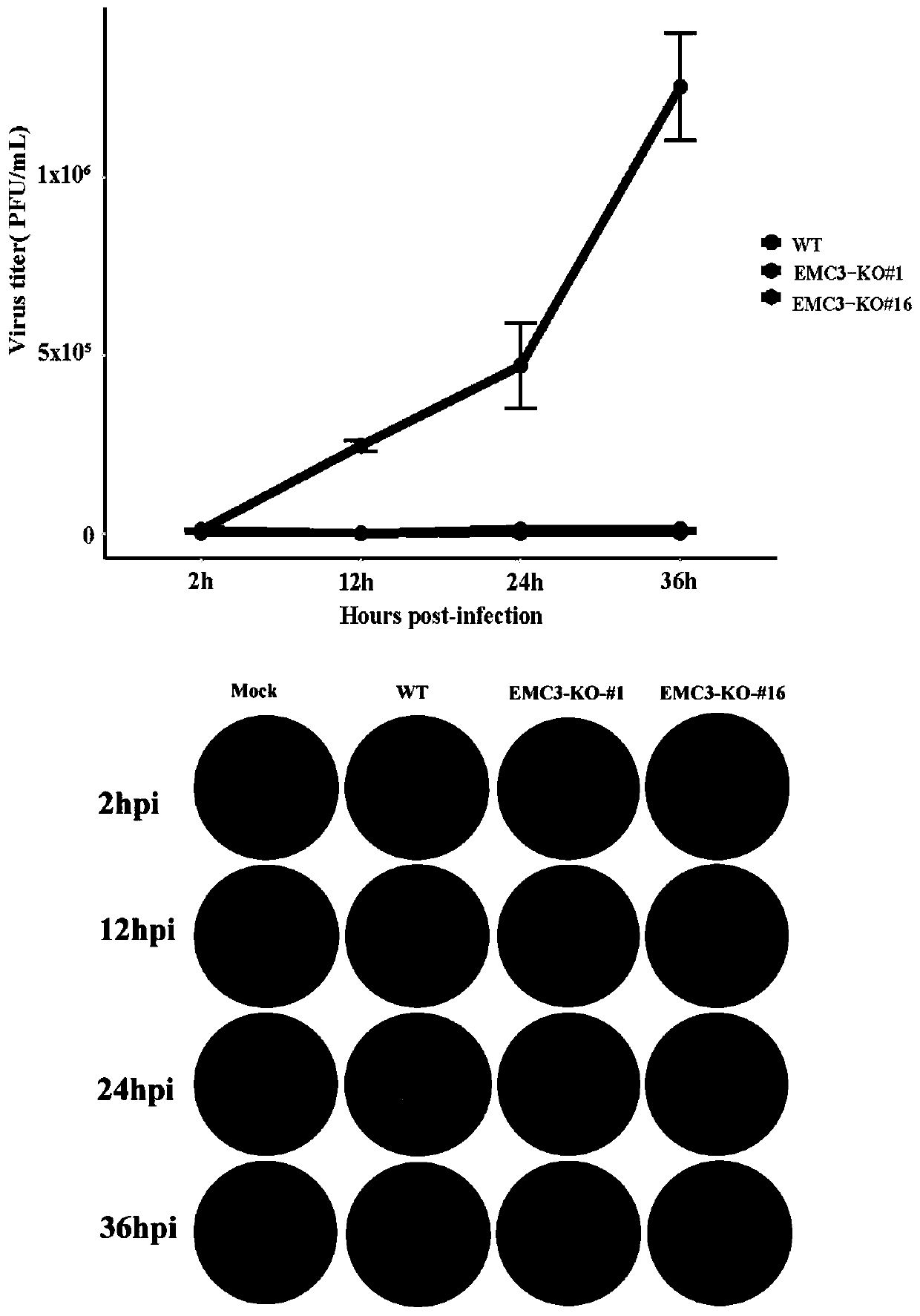
[0055] Example 2: Absolute quantification and plaque experiments found that knocking out the EMC3 gene can significantly inhibit the replication ability of JEV in host cells
[0056] In order to detect whether knocking out the EMC3 gene can inhibit the replication of JEV in PK-15 cells, absolute quantification and plaque assay were used to detect the effects of two monoclonal cell lines, EMC3-KO#1 and EMC3-KO#16, on the replication of JEV. The specific experimental process is as follows:
[0057] First, multiple groups of EMC3-KO#1 and EMC3-KO#16 cells were inoculated at the same time, and PK-15 wild-type cells stably expressing Cas9 were set as the control group. When the confluence reaches about 50%, add the corresponding volume of JEV-RP9 wild-type virus to each well according to MOI=1, shake well and put it back into the cell culture incubator for culture, and change to 2% FBS medium for 2 hours to continue the culture. At different time points such as 12h, 24h, 36h, and ...
Embodiment 3
[0072] Example 3: Using immunofluorescence experiments to find that knocking out the EMC3 gene can significantly inhibit the expression of JEV-encoded protein in host cells
[0073] Furthermore, immunofluorescence assay was used to detect the expression of JEV-encoded gene NS3 at 12 hours after JEV infection of EMC3-KO#1 and EMC3-KO#16 cells. The specific experimental process is as follows:
[0074] Inoculate EMC3-KO#1 and EMC3-KO#16 cells, set PK-15 wild-type cells stably expressing Cas9 as a control, and when the confluence reaches about 90%, add a corresponding volume of JEV-RP9 wild-type cells to each well according to MOI=1 Type virus, after shaking well, put it back into the cell incubator for culture, and change to 2% FBS medium in the 2nd hour and continue to cultivate until the 12th hour. The cells at 12h were fixed with paraformaldehyde PFA, treated with 0.3% Tritonx-100, added blocking solution at room temperature for 1h, NS3 (JEV) primary antibody was incubated ov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com