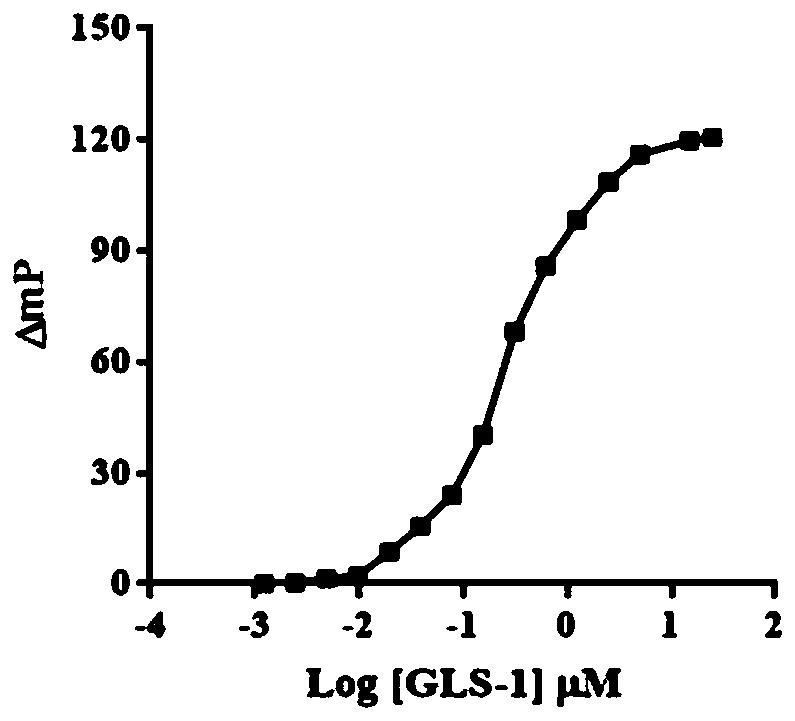
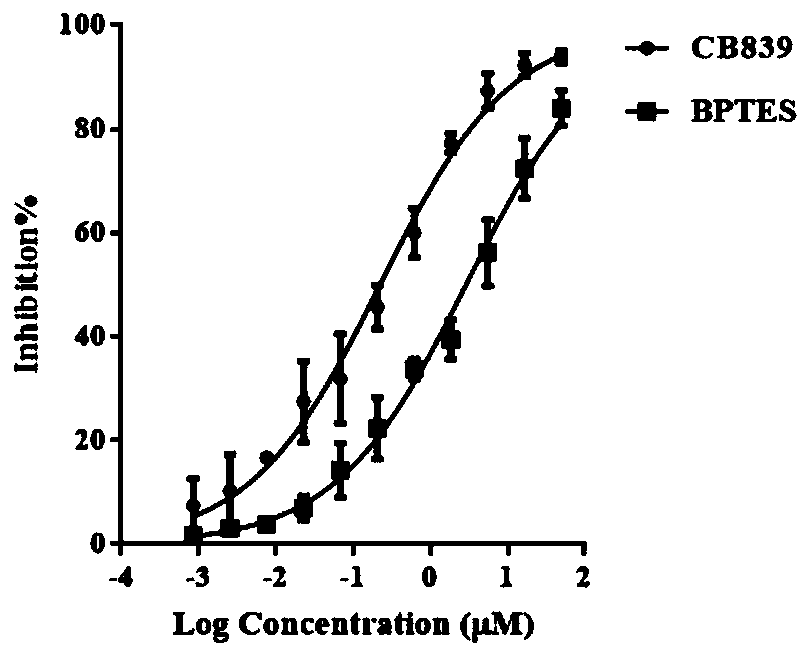
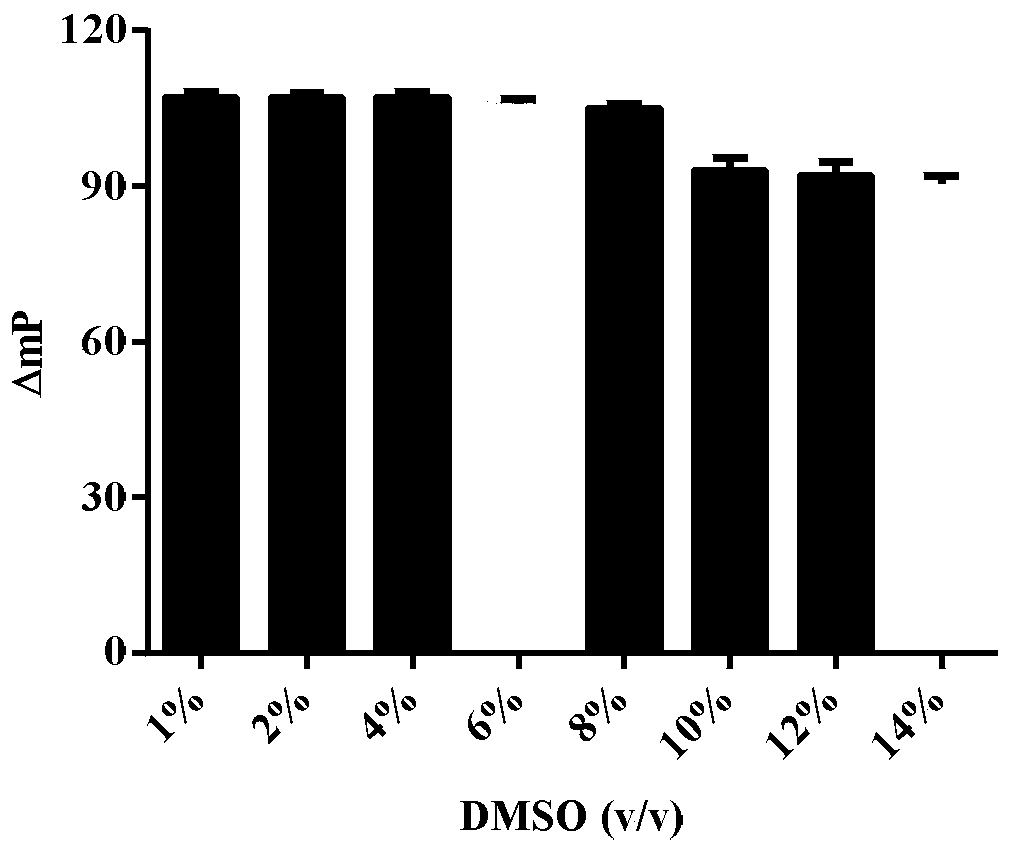
Glutaminase micromolecular fluorescent probe and preparation method and application thereof
A glutaminase, fluorescent probe technology, applied in fluorescence/phosphorescence, chemical instruments and methods, analytical materials, etc., can solve the problems of false positives and false positives in experimental data, and achieve a simple test system with little solvent influence. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Preparation of fluorescent probe Probe 1
[0060]
[0061] 5-(But-3-yn-1-yl)-1,3,4-thiadiazol-2-amine (4)
[0062] Add thiosemicarbazide (1.39 g, 0.015 mol) to the phosphorus oxychloride solution of compound 3 (1.5 g, 0.015 mol), and the reaction mixture was refluxed for 4 hours at 80 ° C. After cooling to room temperature, the reaction mixture was added into 30ml of ice water, adjust the pH to 9 with sodium hydroxide, extract the aqueous layer with ethyl acetate (15ml*3), dry over anhydrous sodium sulfate, filter and concentrate to obtain 1.88g of tan powder, yield 80.2%. 1 H NMR (300MHz, DMSO-d 6 ): δ6.99(s, 2H), 2.95(t, J=7.0Hz, 2H), 2.83(d, J=2.4Hz, 1H), 2.50(dd, J=7.1, 4.6Hz, 2H).HRMS (ESI): m / z, calcd for C 6 h 7 N 3 S[M+H] + , 154.0433; found: 154.0434.
[0063] N-(5-(but-3-yn-1-yl)-1,3,4-thiadiazol-2-yl)-2-(pyridin-2-yl)acetamide (5)
[0064] To compound 4 (3g, 0.02mol) in N,N-dimethylformamide solution, add 2-pyridine acetate hydrochloride (3.74g, 0....
Embodiment 2
[0078] Preparation of fluorescent probe Probe 2
[0079] Scheme 2. Synthesis of probe 2 a
[0080]
[0081] a Reagents and conditions: (a) N-boc-ethylenediamine, trimethylamine, DMF, DCM, rt, 1h, 76%; (b) trifluoroacetic acid, DCM, rt, 10h, 81%; (c) 8, HATU, DIPEA, DMF, 35°C, 1h, 52%.(2-((7-nitrobenzo[1,2,5]oxadiazol-4-yl)amino)ethyl)carbamate tert-butyl ester (12)
[0082] To a solution of compound 11 (0.5 g, 2.51 mmol) in N,N-dimethylformamide was added mono-boc ethylenediamine (0.4 g, 2.51 mmol) and trimethylamine (0.28 g, 2.77 mmol) dissolved in dichloromethane ), the reaction mixture was stirred at room temperature for 1 h, the mixture was added to water (30 ml), the aqueous layer was extracted with ethyl acetate (10 ml * 3), dried over anhydrous sodium sulfate, filtered and concentrated to obtain a solid 0.62 g, yield 76%. 1 H NMR (400MHz, CDCl 3 ): δ8.49(d, J=8.6Hz, 1H), 7.69(s, 1H), 6.18(d, J=8.6Hz, 1H), 5.12(s, 1H), 3.63(s, 4H), 1.48 (s, 9H).HRMS(ESI): m / z, c...
Embodiment 3
[0088] Preparation of fluorescent probe Probe3
[0089] Scheme 3. Synthesis of probe 3 a
[0090]
[0091] N 1 -(2-(3-(3′,6′-dihydroxy-3-oxo-3H-spiro[isobenzofuran-1,9′-xanthene]-6-yl)thioureido)ethyl )-N 5 -(5-(4-(5-(2-phenylacetylamino)-1,3,4-thiadiazol-2-yl)butyl)-1,3,4-thiadiazol-2-yl ) glutaramide (probe 3)
[0092] Please refer to Example 1 for the synthesis method. Yellow solid, yield 36.2%. mp141-143℃. 1 H NMR (300MHz, DMSO-d 6 ): δ12.82(s, 1H), 10.99(s, 1H), 10.00(s, 1H), 9.47(s, 1H), 8.53(d, 2H), 8.37(d, 1H), 8.23(s, 1H), 8.02(d, 1H), 7.60(s, 1H), 7.44(d, 1H), 7.26(d, 1H), 5.34(d, 1H), 4.79-4.71(m, 2H), 3.24(d , 2H), 2.06(s, 3H)ppm; 13 CNMR (75MHz, DMSO-d 6 ): δ178.2, 172.0, 168.3, 164.4, 159.9, 158.5, 155.3, 154.5, 149.5, 145.8, 144.9, 144.5, 138.4, 137.9, 136.1, 133.7, 131.2, 125.4, 123.9, 18, 103.3, 116 57.7, 45.6, 37.67, 35.74, 34.99, 34.70, 28.7, 23.9ppm; HRMS (ESI): m / z, calcd for C 44 h 42 N 9 o 8 S 3 [M+H] + , 920.2313; found: 920.2319. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com