Detection method for hypoglycemic function of marine hypoglycemic health food
A technology of health food and detection method, applied in the direction of measuring devices, instruments, scientific instruments, etc., can solve the problems that synthetic astaxanthin does not have the effect of natural ingredients, the price of natural astaxanthin is high, and the large-scale production and application are restricted. Achieve significant free radical scavenging effect and prevent kidney damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1: Acquisition of astaxanthin high-yield and expression-promoting genes
[0018] The marine red yeast (Rhodotorula benthica) strain was purchased from the China General Microorganism Culture Collection and Management Center, with a preservation number of CGMCC2.5690. Previous studies have proved that the strain has a high astaxanthin production, but it grows in a high-pressure, high-salt environment in the ocean. It is not easy to be cultivated in large-scale industrialization, and cannot be directly applied to the production of industrialized astaxanthin. The above-mentioned problems have not been solved. The present invention explores its molecular mechanism by excavating the high-yield astaxanthin-promoting gene of the marine Rhodotorula yeast, and extracts its genomic DNA for whole-genome sequencing. The genome sequencing was commissioned by Nanjing GenScript Company through the SOLEXA sequencing platform, and the Escherichia coli expression system (E.coli DH...
Embodiment 2
[0020] Example 2: Astaxanthin high-yield expression-promoting gene transfer into cloning vector
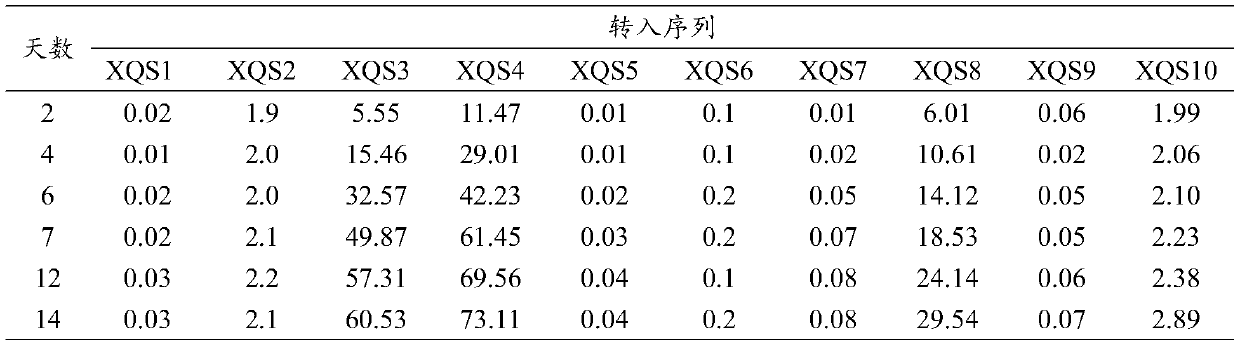
[0021] The nucleotide sequences of XQS1, XQS2, XQS3, XQS4, XQS5, XQS6, XQS7, XQS8, XQS9, and XQS10 were synthesized by Nanjing KingScript Biotechnology Co., Ltd., and the 5' end of the synthesized sequence was also connected with an NcoI restriction site point, the 3' end is also connected with a SwaI restriction site. The synthetic XQS1, XQS2, XQS3, XQS4, XQS5, XQS6, XQS7, XQS8, XQS9, and XQS10 nucleotide sequences were respectively linked into the cloning vector pGEM-T (Promega, Madison, USA, CAT: A3600), and the operation steps were as follows: According to Promega’s product instructions, the recombinant cloning vectors pGEM-XQS01, pGEM-XQS02, pGEM-XQS03, pGEM-XQS04, pGEM-XQS05, pGEM-XQS06, pGEM-XQS07, pGEM-XQS08, pGEM-XQS09, pGEM-XQS10, The vector structure includes: Amp represents the ampicillin resistance gene; f1 represents the replication origin of phage f1; LacZ represen...
Embodiment 3
[0022] Embodiment 3: the preparation of the protoplast of Pichia pastoris
[0023] The protoplasts of Pichia pastoris (Pichia pastoris) were prepared, and the strains of Pichia pastoris (Pichia pastoris) were purchased from the China General Microorganism Culture Collection and Management Center (preservation number CGMCC1.2.5688), and the source of the strain was Fuzhou University Life Science and School of Engineering, the preparation process is as follows: Dilute Pichia pastoris somatic cell culture solution to 1×10 6 cells / ml, add 15ml of EDTA (0.05mol / L) and 1ml of B-mercaptoethanol (015mol / L) to 5ml of diluent, pretreat at 35°C for 15min, then centrifuge at 3500r / min for 10min to collect the bacteria The body was washed twice with hypertonic buffer, then 5ml of 1% cellulase was added to mix well, and then placed in a water bath at 30°C for 60 minutes, then centrifuged at 3500r / min for 10 minutes in a low-temperature centrifuge to collect the cells, and then used a high ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com