Screening of schistosoma japonicum W chromosome specific gene and application thereof in cercaria sex identification
A technology for specific genes and schistosomiasis, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of screening out nucleic acid sequences or genes, etc., to improve standardization and homogenization, and has a wide range of applications. highly reproducible results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
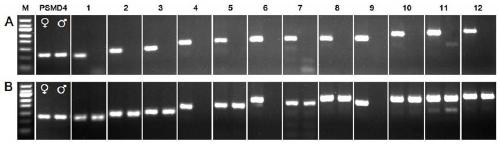
[0042] Example 1 Schistosoma japonicum W chromosome-specific gene screening
[0043] (1) 100 female-specific expression genes were preliminarily screened out based on the ratio of fluorescent signals expressed by male and female adults, and 12 candidate genes whose gene sequences were better compared to the Schistosoma japonicum genome were screened out through bioinformatics analysis.
[0044] (2) Design of PCR primers: According to the gene sequence, gene-specific primers were designed using PrimerPremier 5.0 software.
[0045] Table 1 Primer information of 12 female specific expression genes of Schistosoma japonicum
[0046]
[0047] (3) PCR screening of Schistosoma japonicum W chromosome-specific genes: the cDNA and genomic DNA of male and female adults of Schistosoma japonicum were respectively used as templates, the housekeeping gene PSMD4 was used as an internal reference gene, and the upstream primer was 5'-CCTCACCAACAATTTCCACATCT-3' (SEQ ID NO. 10), the downstream...
Embodiment 2
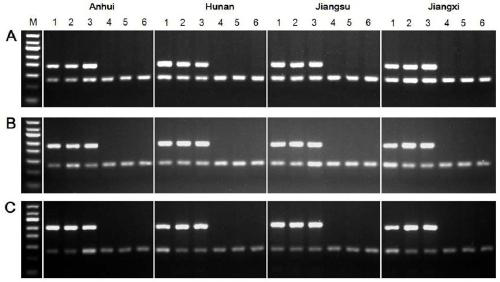
[0051] Example 2 Verification of Schistosoma japonicum W chromosome-specific genes in different geographical strains
[0052] (1) Genomic DNA extraction: collect the male and female adults of Schistosoma japonicum in Anhui, Hunan, Jiangxi and Jiangsu provinces in China, and use the Genomic DNA Extraction Kit (QIAGEN Company) to extract the genomic DNA of the male and female adults respectively (the female and male worms were biologically extracted three times respectively). duplicates) to prepare PCR templates.
[0053] (2) Using the genomic DNA of male and female adults of Schistosoma japonicum in different provinces as templates and PSMD4 as an internal reference gene, double-PCR detection was carried out using the primers of SjF4, SjF6 and SjF9 specific genes of Schistosoma japonicum W chromosome, respectively. The PCR reaction system is as follows:
[0054]
[0055] The PCR amplification procedure is the same as in Example 1.
[0056] (3) Detect the double PCR product...
Embodiment 3
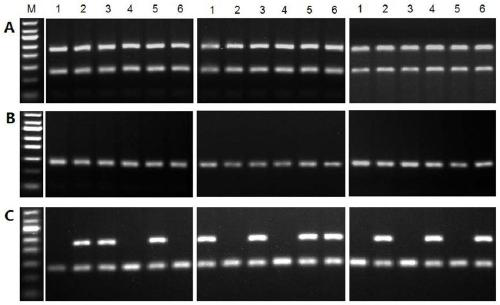
[0057] Example 3 Preparation of positive snails infected by single-sex and mixed-sex Schistosoma japonicum
[0058] (1) Negative snail screening: Oncomelania were collected in non-schistosomiasis-endemic areas, and further verified as non-schistosomiasis-infected negative snails by the light escape method.
[0059] (2) Preparation of positive snails artificially infected: a single snail was infected by a single miracidia or a mixed infection of multiple miracidia was used to infect a snail, and the snails were cultured in a light incubator for 3 months after infection.
[0060] (3) Identification of cercariae sex by animal infection experiment: put one artificially infected snail into each test tube and number them, and release the cercariae in a single positive snail by light-escape method, and pick about 60 cercariae from each snail and pass them through the abdominal patch Mice were infected one-on-one, and the infected mice were fed for 5 weeks and then dissected to obtain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com