Method using blue algae to synthesize phloretin
A phloretin and cyanobacteria technology, applied in microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of inability to achieve large-scale production, low phloretin efficiency, and chemical waste liquid discharge, etc. The effect of industrial transformation, simple operation and production cost reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
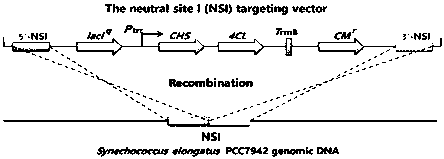
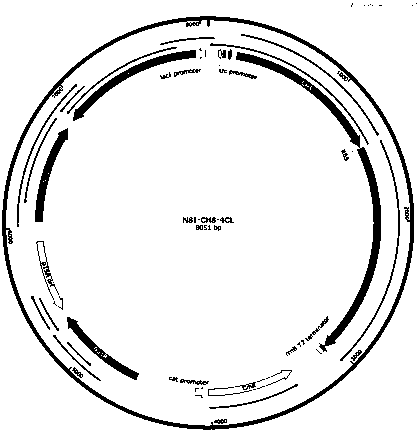
[0035] This embodiment provides the vector plasmid construction process: according to the method provided in the present invention Synechococcus elongatus The NSI gene sequence of PCC7942 (Syn7942 for short), according to the attached figure 2 The plasmid map in the construction of the integration vector NSI, and then according to the amino acid sequence of 4CL and CHS provided in the present invention, the nucleic acid sequence of the corresponding gene is designed according to the codon preference of Syn7942 cells, and the gene 4CL and CHS are sent to companies with gene synthesis capabilities on the market Synthesize, design a homology arm with a size of about 20bp, follow the attached image 3 In the plasmid map, the two genes were constructed on the integration vector NSI using a one-step cloning kit.
[0036] NSI as Synechococcus provided by the present invention Synechococcus elongatus A gene sequence on the PCC7942 genome, the role of NSI is that changes in this gen...
Embodiment 2
[0039] This embodiment provides the recombinant transformation process:
[0040] 1) Take 2 mL of wild-type Syn7942 with a growth density of OD730 of 0.8-1.2 in a centrifuge tube, then centrifuge at 10,000 rpm for 2 min, and remove the supernatant;
[0041] 2) Mix the precipitate in the previous step with 1mL 10mM sodium chloride solution, then centrifuge at 10000rpm for 2min, and remove the supernatant;
[0042] 3) Mix the precipitate in the previous step with 1 mL of sterilized anti-BG11-free liquid medium, then centrifuge at 10,000 rpm for 2 min, and remove the supernatant;
[0043] 4) Mix the precipitate in the previous step with 100 uL of sterilized anti-BG11-free liquid medium, and then add 200 ng of plasmid DNA to it. Seal the centrifuge tube thoroughly with tinfoil (protect from light), and then incubate in a shaker at 100 rpm at 30°C for 10 hours;
[0044] 5) Apply all the cyanobacteria in the centrifuge tube in the previous step to the BG11 solid medium containing 2...
Embodiment 3
[0047] This embodiment provides the screening confirmation of transgenic cyanobacteria and the detection process of the product:
[0048] Pick about 10 monoclonal cyanobacterial plaques grown on BG11 solid medium containing 25ug / mL chloramphenicol, insert them into 5mL 25ug / mL chloramphenicol BG11 liquid medium for culture, and extract For the genome, verify the two target genes of CHS and 4CL by PCR with the designed primers, and send the products to sequence by PCR to determine whether the gene sequence is correct; insert the Syn7942 cyanobacteria that have been verified to have the correct transgenic CHS gene and 4CL gene into 100mL BG11 liquid medium Medium light culture, when the OD730 was 1, the substrate p-hydroxyphenylpropionic acid was added, and IPTG was added to induce the expression of CHS and 4CL genes, and then the transgenic cyanobacteria were cultured for 1 day, 3 days, and 5 days after induction, respectively, and used The high-pressure crusher breaks the cyan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com