Method for knocking out CRISPR/Cas9 mediated sheep FGF5 gene and integrating MTNR1A gene at fixed point
A gene and gene editing technology, applied in the field of genetic engineering, can solve problems such as slow progress in sheep fecundity and low fecundity of high-quality meat varieties, and achieve the effect of avoiding abnormal gene function and biological safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Construction of CRISPR / Cas9 Targeting Vector
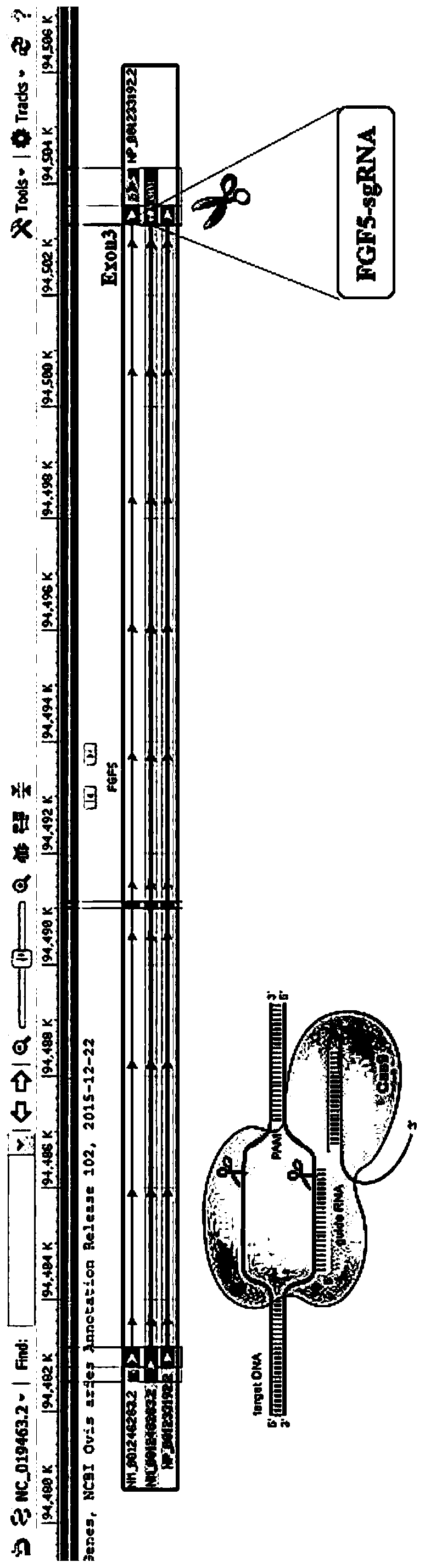
[0046] The targeting site is located in the third exon of the sheep FGF5 gene (NM_001246263.2), and the corresponding targeting site is as follows figure 1 As shown, wherein, the nucleotide sequence identifying the target in the sgRNA sequence is shown in SEQ ID NO: 1, and the DNA sequence encoding the above-mentioned sequence is shown in SEQ ID NO: 2. According to the above target sequence, the corresponding primer sequence was designed and synthesized by Sangon Biotechnology Co., Ltd., and the purification method was HPLC. The specific sequence is shown in Table 1.
[0047] Table 1 Goat FGF5 gene targeting sequence primers
[0048] Nucleotide name
sequence (5'-3')
Ovis aries-FGF5-sgRNA-F
caccgAGGTTCCCCTTTCCGCACCT
Ovis aries-FGF5-sgRNA-R
aaacAGGTGCGGAAAGGGGAACCTc
[0049] Formation of Olio nucleic acid sequence: Dilute primers to 100 μM, phosphorylation anneal, system: Ov...
Embodiment 2
[0051] The construction of embodiment 2 donor vector
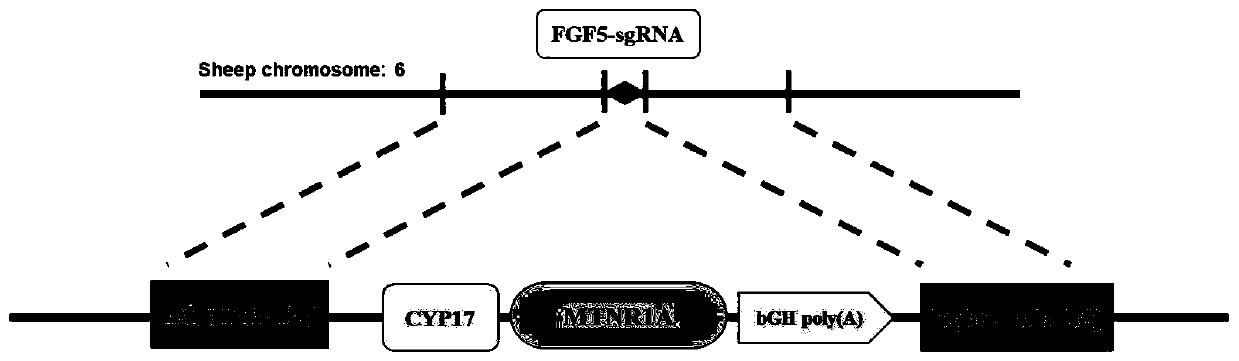
[0052] Such as image 3 As shown, in order to insert MTNR1A into the FGF5 target site, a gene locus with a homology arm needs to be inserted into the target site by means of Homology directed repair (HDR). Since the construction of the donor vector requires cloning four long fragments into a destination vector, coupled with the uncertainty of the restriction site, it is difficult to ensure that each fragment can be spliced properly, so Gibson Assembly based on homologous recombination was used , which can stitch together more long fragments efficiently and seamlessly without restriction of enzyme cutting sites.
[0053] The construction process of the donor vector is as follows: Figure 4 As shown, the upstream homology arm 5-HA, CYP17-promoter, MTNR1A(MT1)-bGH poly(A) signal, and the downstream homology arm 3-HA need to be ligated into the backbone vector pUC57-Amp, where the upper, The downstream homology arm and CY...
Embodiment 3
[0061] Example 3 Cell Transfection
[0062] 1. Prepare fetal fibroblasts by conventional methods.
[0063] 2. Adopt nuclear transfer method.
[0064] with nucleofection-Amaxa TM Basic Nucleofector TM Kit for Primary Mammalian Fibroblasts (Lonza), using Nucleofector TM The program V-024 of 2b was used for transfection, 10 micrograms of the donor vector and 10 micrograms of the targeting vector. For the transfection steps, please refer to the kit instructions. For transfection see Image 6 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com