Composite amplification kit for 47 human autosome and Y chromosome loci and application thereof
An autosome and Y-chromosome technology, applied in the field of molecular genetics and biological detection, can solve the problems of poor kit compatibility and limited number of STR loci
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 147
[0069] Example 1 Development of multiple amplification kit for 47 human autosomes and Y chromosome loci
[0070] 1. Design of multiple amplification primers for 47 human autosomal and Y chromosome loci
[0071] For the composite amplification system, especially the composite amplification system of 47 loci described in the present invention, the requirements for the specificity of the primer sequence, the secondary structure, the stability of binding to the target sequence, and the amplification efficiency are extremely high. , after continuous amplification-optimization-amplification cycle experiments, 47 pairs of primers for specifically and efficiently amplifying 47 loci were obtained, the specific sequences of which are shown in Table 1:
[0072] Table 1 Amplification primers of 47 loci
[0073]
[0074]
[0075] The above primers were fluorescently labeled with different colors according to the following grouping methods corresponding to the loci: the first group, ...
Embodiment 24
[0082] Example 2 Establishment of multiple amplification system and program for 47 chromosomal loci
[0083] The annealing temperature and cycle number were optimized for the amplification primers of 47 loci, and the optimal reaction program was obtained: 95°C for 2 minutes; 94°C for 5 seconds; 60°C for 45 seconds; 72°C for 45 seconds Minutes, this step runs 30 cycles; 60°C for 20 minutes; 4-10°C for heat preservation.
Embodiment 3
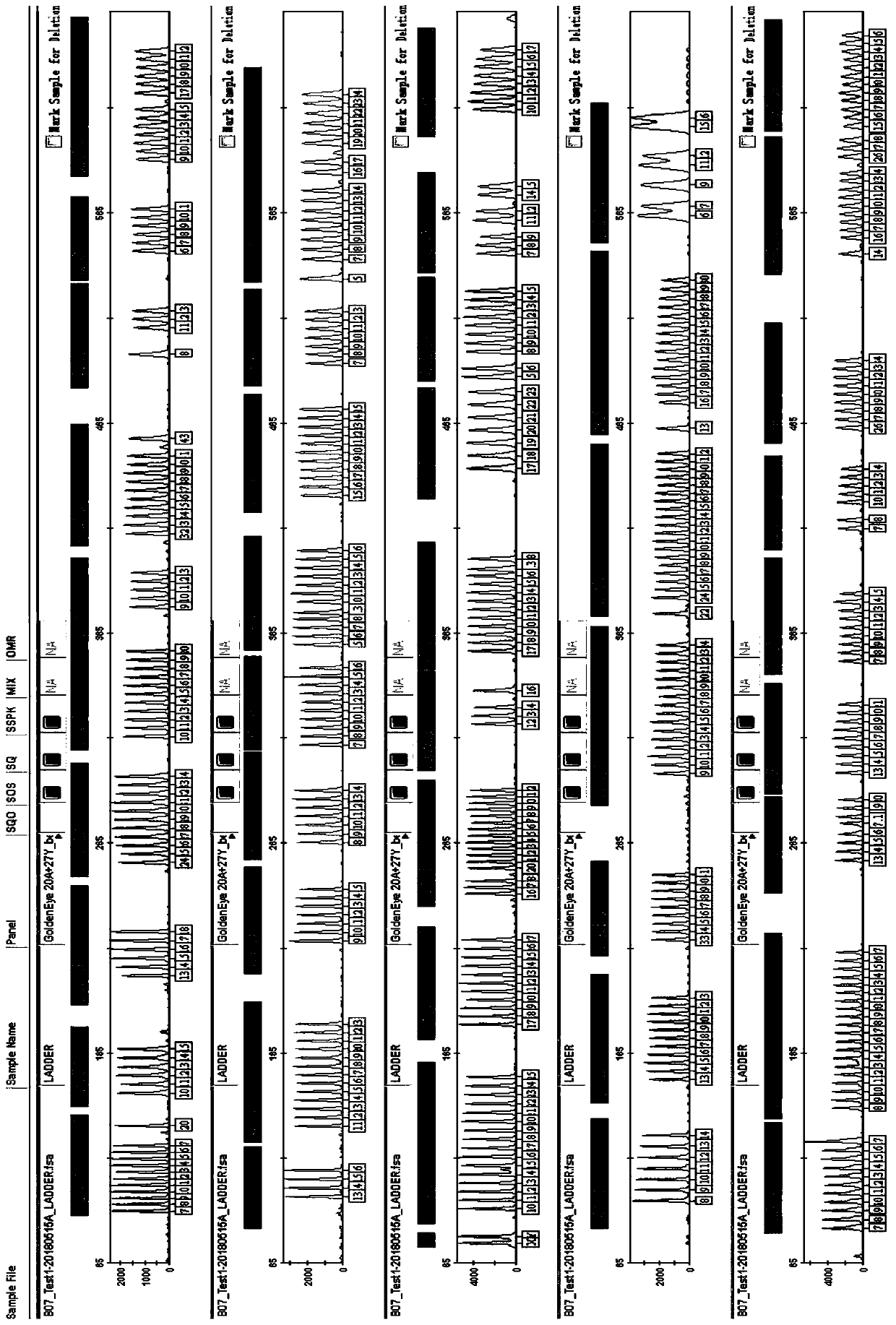
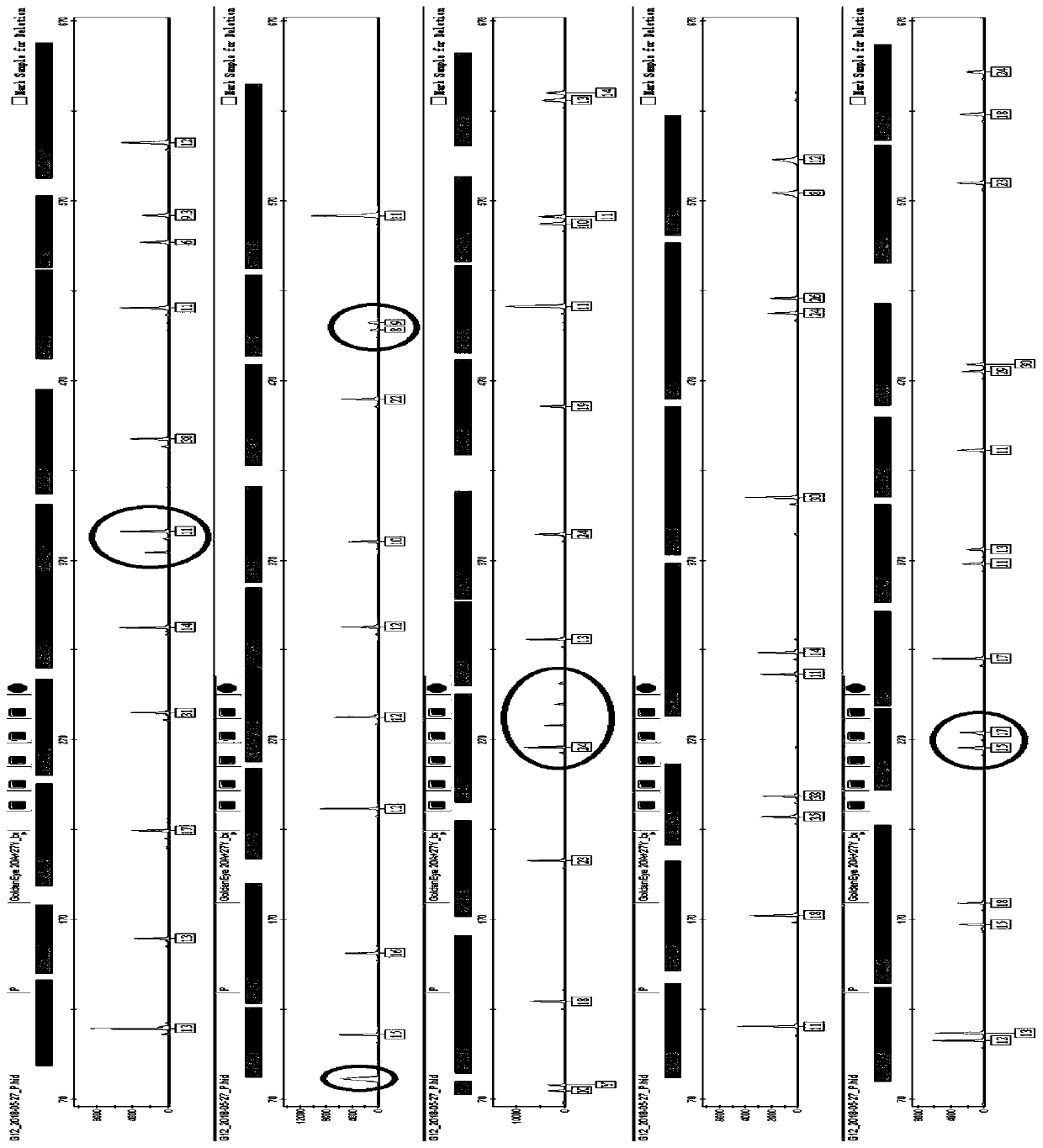
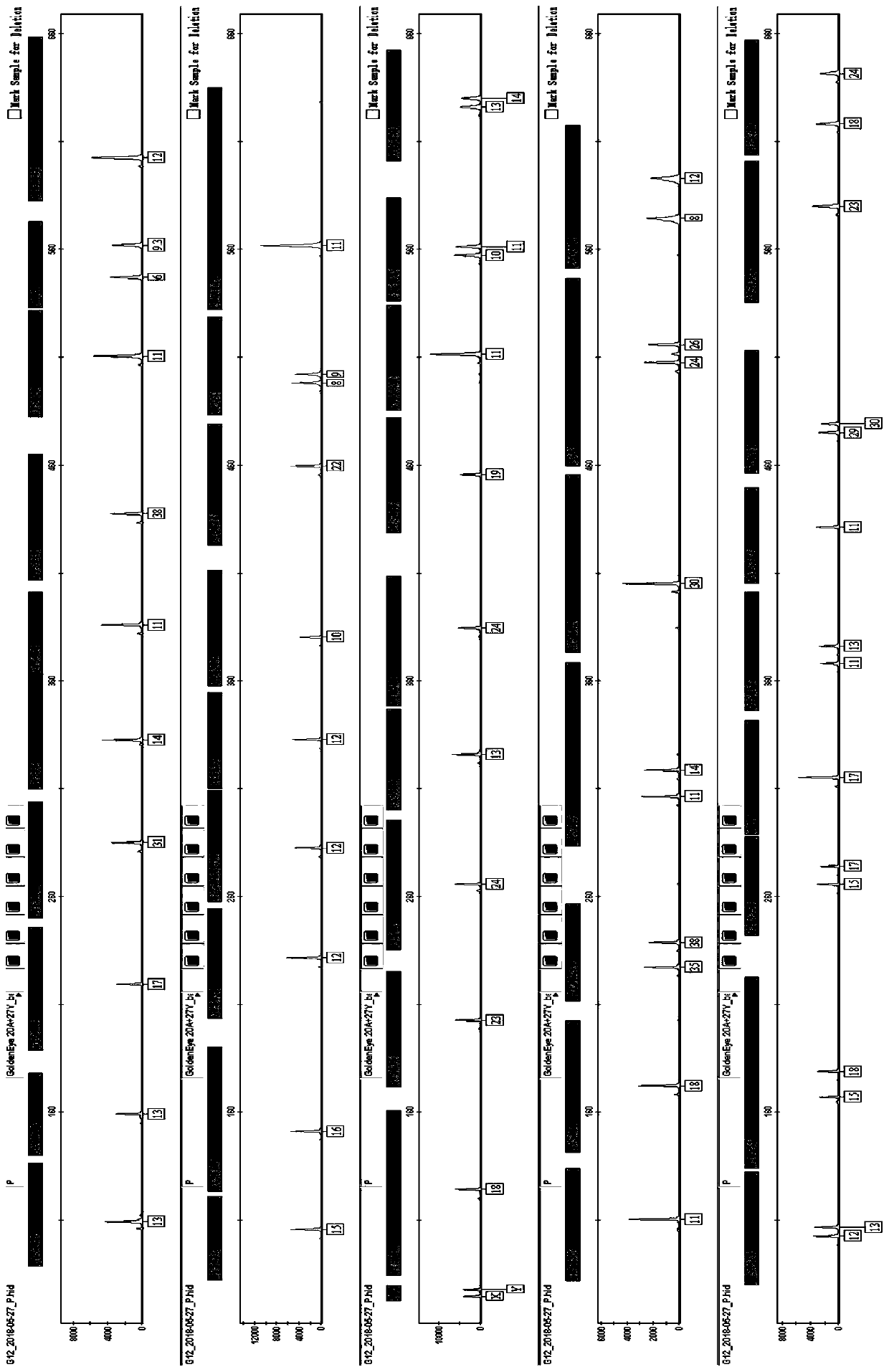
[0084] Example 3 Genotyping of 9948 Cell Line Using Multiplex Amplification Kit
[0085] The kit provided by the invention is used to carry out multiple amplification of 47 human autosome and Y chromosome loci on the 9948 cell line. The template DNA derived from 9948 cell line was extracted by Chelex 100 method. The amplification reaction was carried out on the ABI 9700 thermal cycler, the electrophoresis and detection were carried out on the ABI 3130 genetic analyzer, and the data analysis was performed using GeneMapperIDX v1.2 software. Reagent materials such as the allelic ladder (ladder) used can be obtained from commercial sources and are conventional materials commonly used by those skilled in the art.
[0086] 1. Use the chelex 100 method to extract DNA (for specific methods refer to "Forensic DNA Protocol", HumanPress, 1998):
[0087] (1) Take 3 μl of cultured 9948 cell line in a 500 μl centrifuge tube.
[0088] (2) Shake and mix the chelex solution to make the chelex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com