Hammerhead ribozyme screened in vitro and capable of shearing RNA
A ribozyme, cell technology, applied in the field of RNA molecules, can solve the problem of insignificant effect and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1. In vivo screening of ribozymes that catalyze intermolecular cleavage based on toxin reporter genes
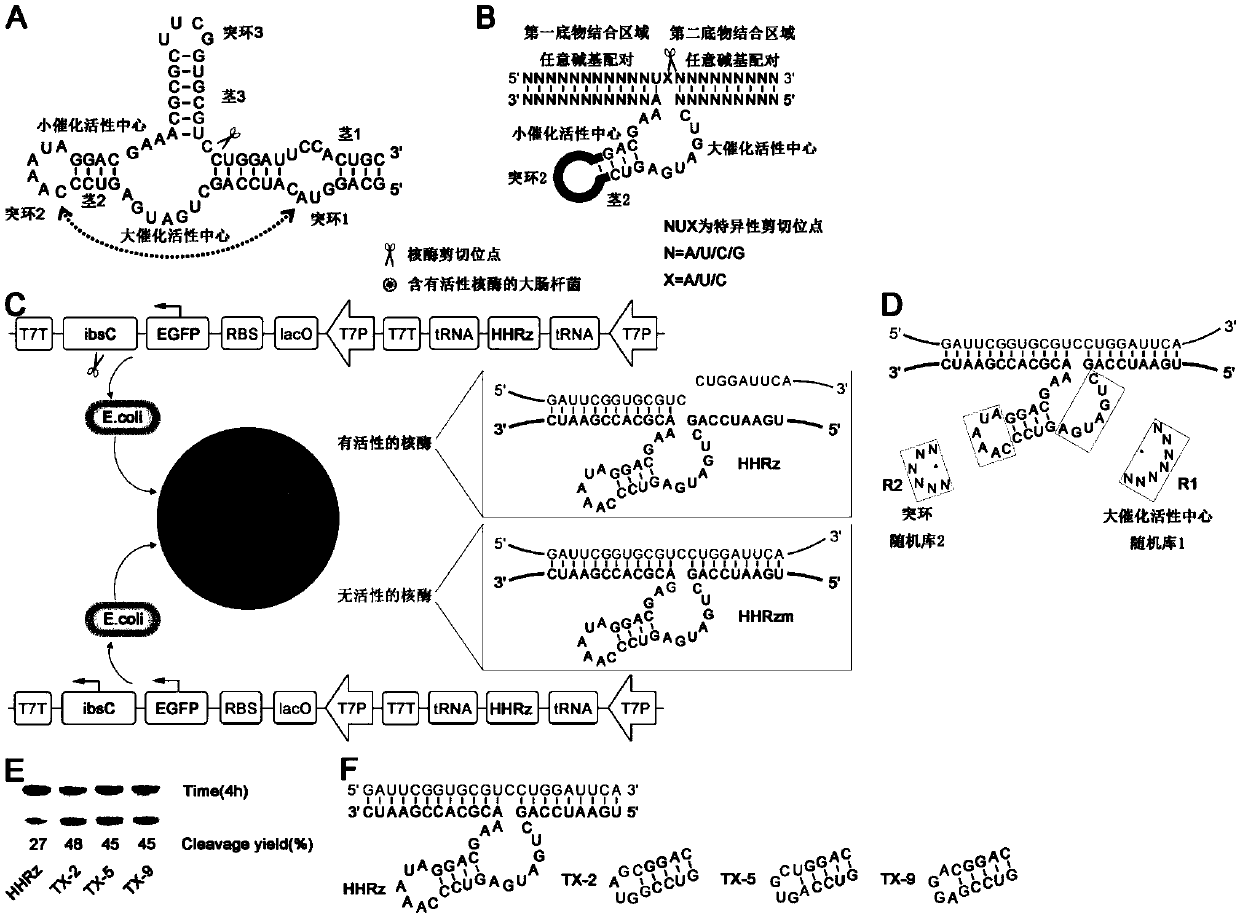
[0030] The naturally occurring hammerhead ribozyme ( figure 1 A) is a ribozyme that catalyzes intramolecular cleavage. There are two highly conserved catalytic active centers (large catalytic active center and small catalytic active center), which consist of three stem-loop structures (stem 1, protruding loop 1, stem 2, Protruding loop 2, stem 3, protruding loop 3) are connected, GUC is one of the ribozyme cleavage recognition sites, and the cleavage position is after GUC ( figure 1 A scissors shown). The efficient cleavage of this ribozyme in cells depends on the interaction between protruding loop 1 and protruding loop 2. When the ribozyme is cleaved at the protruding loop 3, an intermolecular cleavage ribozyme can be formed, but the interaction between the protruding loop 1 and the protruding loop 2 still needs to be retained-the cleavage efficiency of th...
Embodiment 2
[0046] Example 2. The ribozyme obtained by screening down-regulates the expression of red fluorescent protein and Spinach in Escherichia coli.
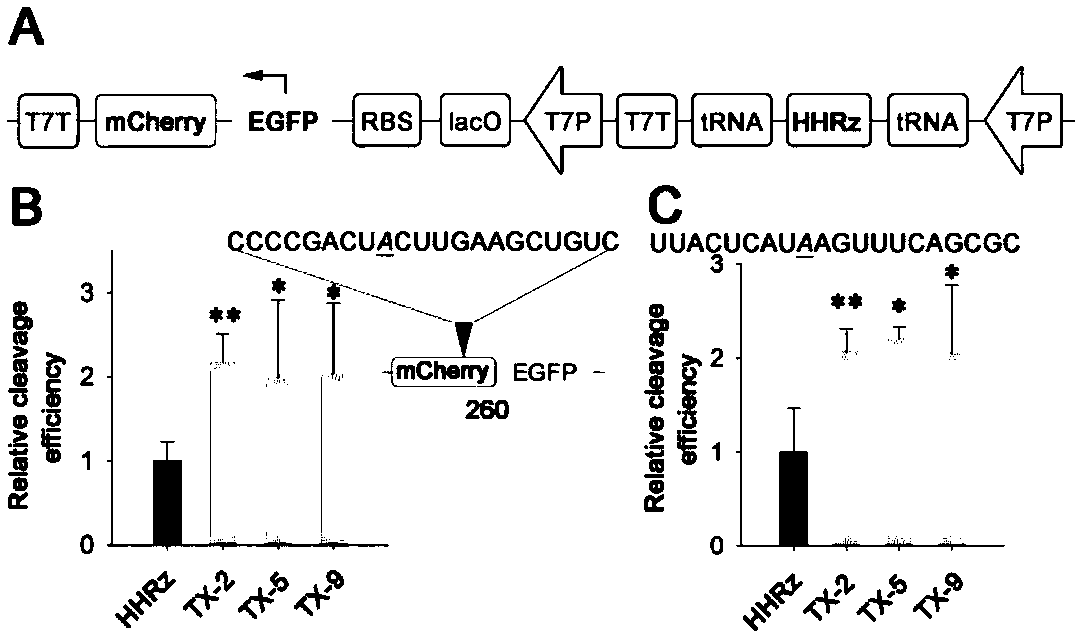
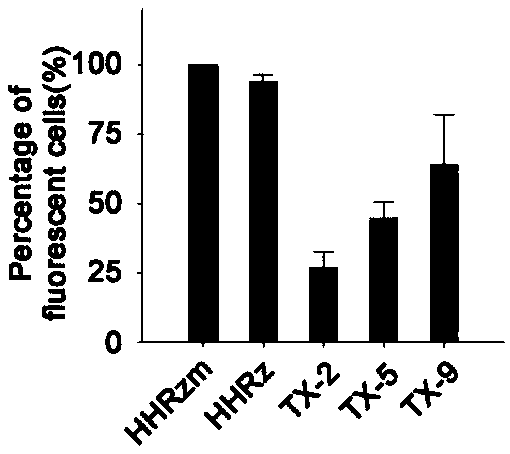
[0047] The ribozyme screened in vivo has two substrate-binding domains (the first substrate-binding domain and the second substrate-binding domain), located on both sides of the catalytic core domain, and the two substrate-binding domains are respectively linked to the preselected RNA The sequences of the substrates are complementary, while the core catalytic domain remains unchanged. In this example, red fluorescent protein RNA (SEQ ID NO 10) and a sequence of Spinach (SEQ ID NO 11) were selected as the target RNA, and the working ability of the preferred ribozyme obtained through screening in prokaryotic cells was investigated. The corresponding plasmid constructs are as figure 2 Shown in A, other conditions are with reference to embodiment 1.
[0048] The red fluorescent protein mCherry was constructed downstream of the green fl...
Embodiment 3
[0050] Example 3. The ribozyme obtained by screening down-regulates the expression of red fluorescent protein in human cancer cell line (Hela).
[0051] The preferred ribozymes obtained from the screening show better cutting efficiency for various genes in Escherichia coli, a prokaryotic cell. In this example, the ability of the ribozyme to down-regulate the red fluorescent protein (SEQ ID NO 10) expressed in a eukaryotic human cell line was tested. The ribozyme obtained by screening was constructed on the eukaryotic expression vector pmCherry by restriction endonucleases SalI and HindIII, and the recombinant ribozyme-containing vector was extracted with an endotoxin-free plasmid extraction kit to obtain a plasmid. 5 micrograms of the recombinant vector used 2000 (Liposome 2000) transfected Hela cells that were plated on a 6-well plate one day in advance. 24 hours after transfection, single cell suspension was obtained by trypsinization, and the red fluorescence intensity o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com