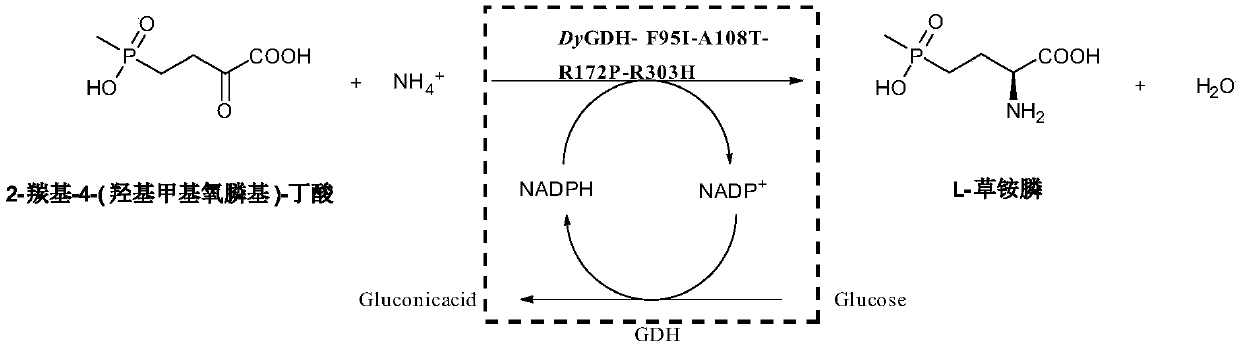
Amino acid dehydrogenase mutant and application of mutant in synthesizing L-glufosinate
An amino acid and dehydrogenase technology, which is applied to amino acid dehydrogenase mutants and its application in the synthesis of L-glufosinate-ammonium, can solve the problems of low substrate concentration and low asymmetric amination reduction activity, and achieve The effect of shortened reaction time and good industrial application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Construction and screening of amino acid dehydrogenase mutant library
[0034] The amino acid dehydrogenase gene (nucleotide sequence shown in SEQ ID No.1, amino acid sequence shown in SEQ ID No.2) cloned from Pseudomonas monteiliiWP_060477601.1 was constructed expression vector pETDEut- dygdh was transformed into Escherichia coli to obtain the starting strain E.coli BL21(DE3) / pETDEut-dygdh.
[0035] The preparation of the amino acid dehydrogenase mutant library was achieved through 4 rounds of site-directed saturation mutagenesis. The primers were designed as shown in Table 1. In the first round, the vector pETDEut-dygdh was used as a template, and F95F and F95R in Table 1 were used as primers, and saturation mutation PCR was carried out. , the 95th phenylalanine in the amino acid sequence of amino acid dehydrogenase shown in SEQID No.2 was mutated into the remaining 19 amino acids, and transformed, plated, and the amino acid dehydrogenase mutant pDyGDH-F95L...
Embodiment 2
[0039] Example 2: Induced expression of amino acid dehydrogenase parent, mutant and glucose dehydrogenase
[0040] Glucose dehydrogenase gene esgdh (nucleotide sequence is shown in SEQ ID No.3, amino acid sequence is shown in SEQ ID No.4) cloned from Exiguobacterium sibiricum, and connected to pET-28b(+) by double restriction enzymes On the vector, the recombinant plasmid was introduced into Escherichia coli E.coli BL21(DE3) to obtain the recombinant glucose dehydrogenase strain E.coliBL21(DE3) / pET28b-esgdh.
[0041] The starting strain E.coli BL21(DE3) / pETDEut-dygdh and the amino acid dehydrogenase mutant strain in Example 1 were respectively inoculated into LB liquid medium containing ampicillin at a final concentration of 50 μg / mL, cultured at 37°C for 8 hours, Inoculate the inoculum with a volume concentration of 2% into fresh LB liquid medium containing ampicillin at a final concentration of 50 μg / mL, culture at 37°C and 180 rpm for 1.5 hours, and then add a final concent...
Embodiment 3
[0044] Example 3: Mutant library screening
[0045] Mix the wet cells of the mutant strain induced and expressed in Example 2 and the wet cells of glucose dehydrogenase at a mass ratio of 3:1, add the amount of 50 g / L of the total amount of cells into pH 7.4, and resuspend in 100 mM phosphate buffer , ultrasonic crushing on ice-water mixture for 15 minutes, ultrasonic crushing conditions: power 400W, crushing for 1 second, pausing for 5 seconds, to obtain the crude enzyme solution of the mutant strain. Under the same conditions, replace the wet cells of the mutant strain with the starting strain E.coli BL21(DE3) / pET2Deut-dygdh to prepare the crude enzyme solution of the starting strain.
[0046] The crude enzyme solution of the mutant strain or the crude enzyme solution of the original strain was used as a catalyst, 2-carbonyl-4-(hydroxymethylphosphinyl)-butyric acid was used as a substrate, glucose was used as an auxiliary substrate, and no exogenous NADPH was added. Or NADP...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com