Detection kit and method for DNA methylation
A detection method and methylation technology, applied in the field of analysis, can solve the problems of quantitative error of absolute value of methylation level, no cost-benefit advantage, inability to detect methylation levels in regions other than CpG islands, etc., and achieve increased annealing. effect of temperature
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0119] Example 1 Quantitative method for DNA methylation at CpG sites
[0120] DNA isolation and bisulfite conversion
[0121] Using the QIAamp DNA Mini Kit (Qiagen, 51306) and the EZ DNA Methylation Kit (ZymoResearch, D5002), the genomic DNA in the above samples was extracted and modified with bisulfite according to the instructions [7,14].
[0122] QASM detection
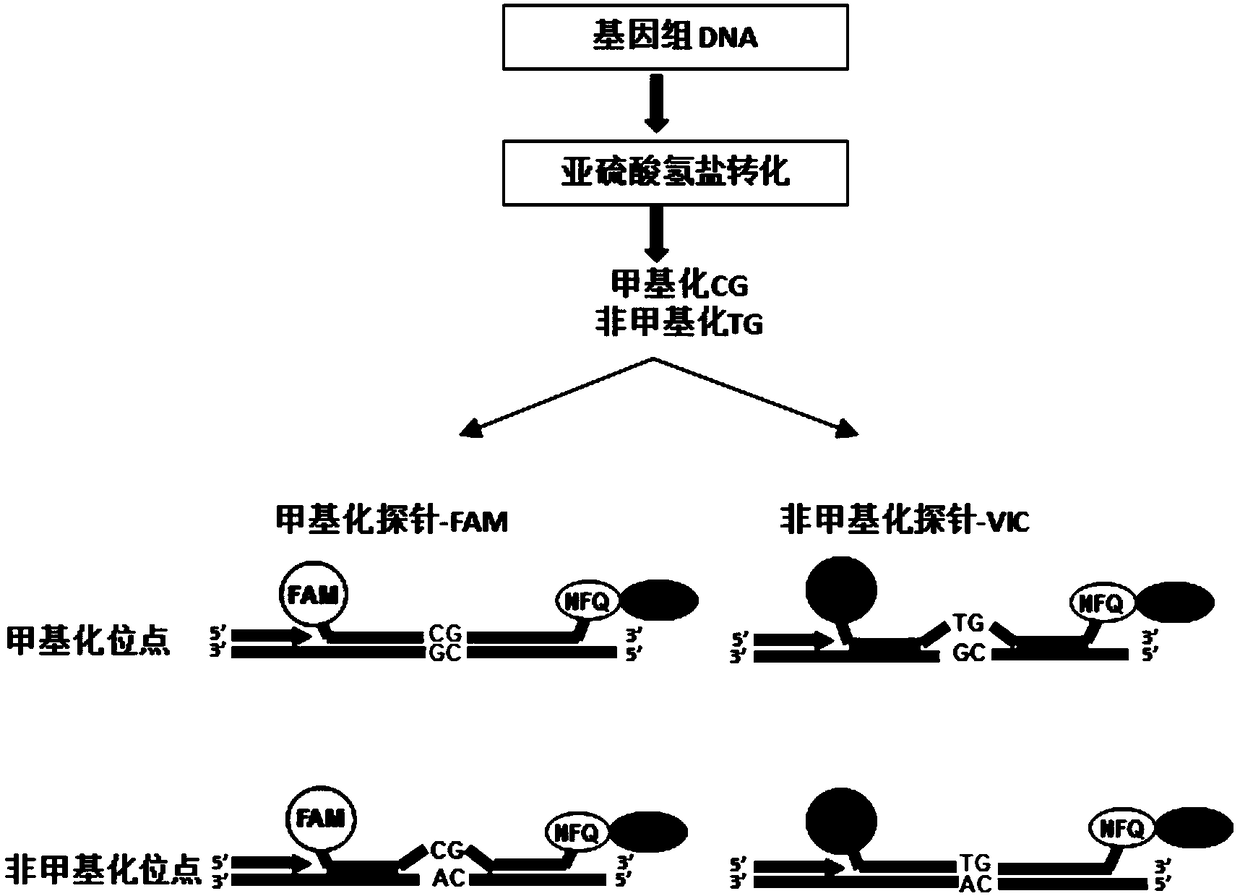
[0123] After bisulfite conversion, real-time fluorescent quantitative PCR was performed to amplify the genomic DNA. Briefly, bisulfite-converted genomic DNA is amplified using primers and a pair of oligonucleotide probes covering the CpG sites to be tested, each oligonucleotide probe linked to a fluorescent reporter at the 5' end Dye 6FAM or VIC (specifically binding to CG sequence and TG sequence, respectively), 3' end coupling quencher-MGB group (MGB-NFQ) ( figure 1 ). Upon DNA extension, the 5' to 3' exonuclease activity of Taq DNA polymerase cleaves the probe and releases the reporter gene, whose fluoresce...
Embodiment 2
[0151] Example 2 Bisulfite Pyrosequencing Comparative Evaluation of Quantitative Accuracy
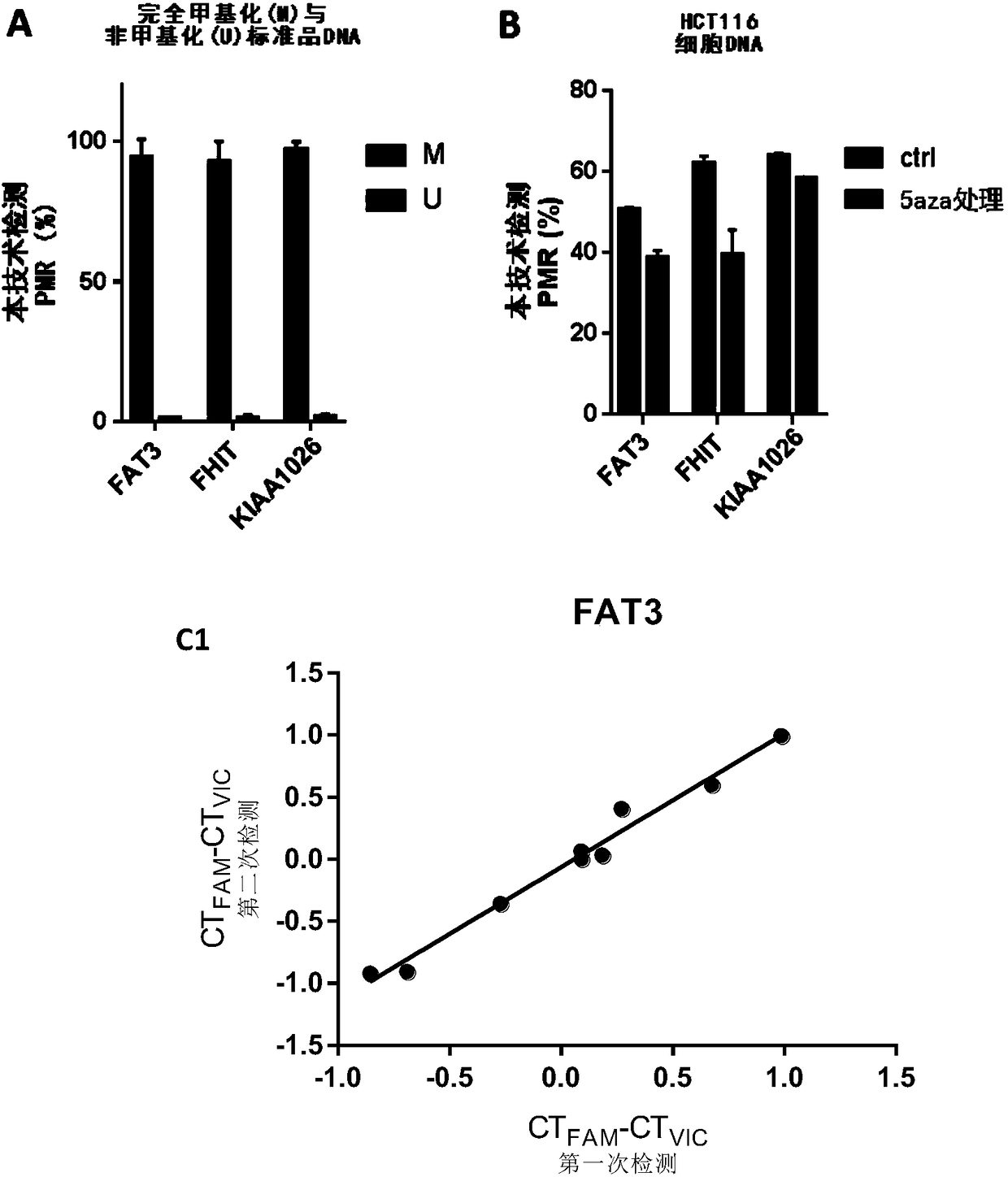
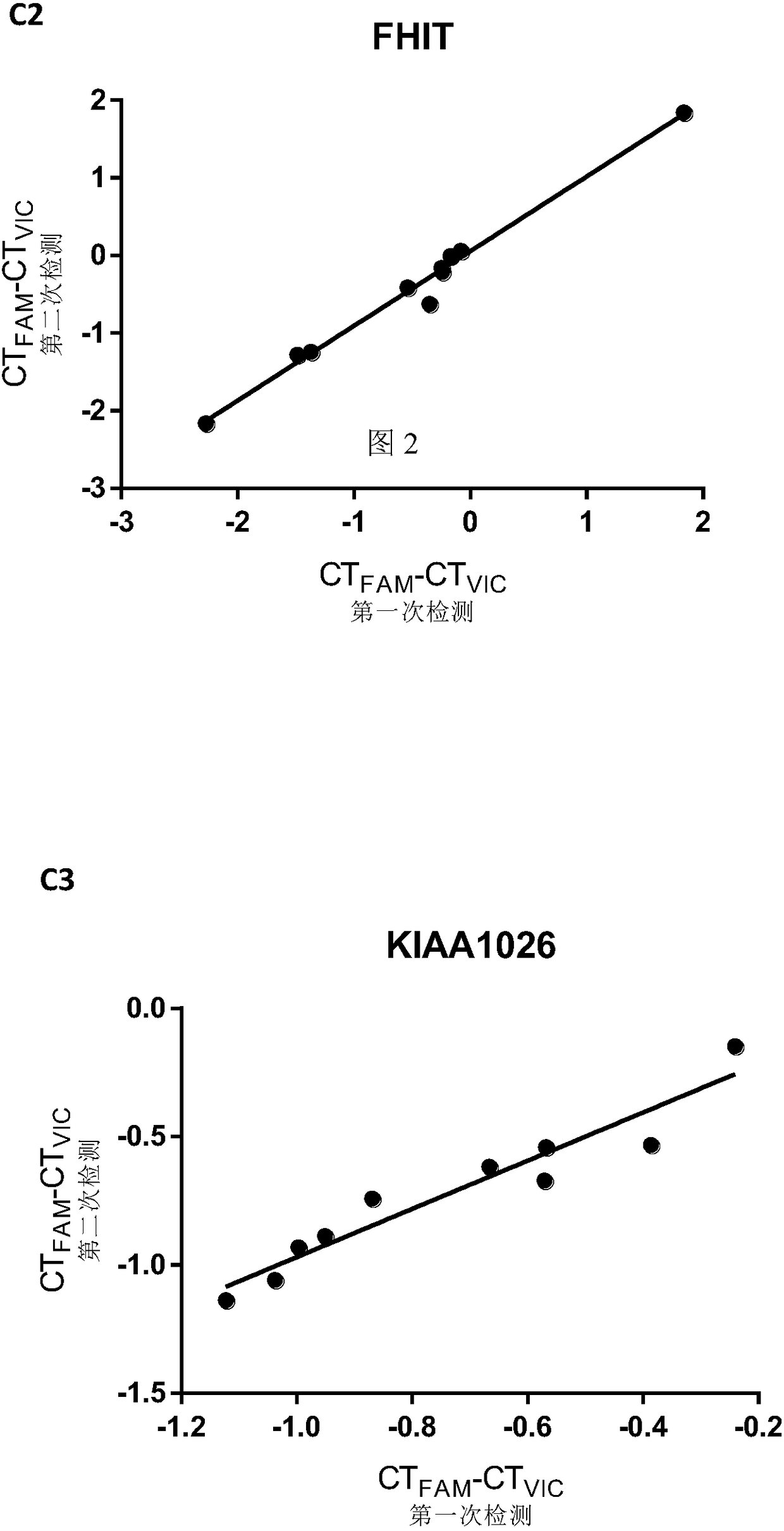
[0152] Using pyrosequencing as a reference, the accuracy of the methylation quantification method of the present invention was further tested. We used pyrosequencing and this technology to detect the methylation levels of the above three CpG sites in 10 colorectal cancer tissues (Table 1). The primers for pyrosequencing are listed in Table 2. The PMR measured by the present invention is linearly correlated with the methylation percentage obtained by pyrosequencing ( image 3 ; FAT3 was 0.9690, FHIT was 0.9954, KIAA1026 was 0.8755, all P<0.001). Not only that, there is a striking agreement between PMR and Pyrosequencing in terms of percent methylation. This is more precise than traditional MethyLight reported previously [11,12]. Therefore, the present technique has the same quantitative accuracy as bisulfite pyrosequencing, but is simpler, more accessible, and less expensive.
Embodiment 3
[0153] Example 3 Comparison of repeatability with techniques using Alu-C4 reaction correction
[0154] In the past, all qPCR-based detection techniques required internal reference genes for quantification. Traditional MethylLight widely uses AluC4 as a control reaction to assess and correct the total amount of starting DNA template [8,11,12]. This high-copy control amplicon is less susceptible to cancer-associated gene alterations than other single-copy control genes. Unfortunately, the effects of genome-wide copy number variation and mutations on AluC4 stability still cannot be completely avoided. The advantage of the present invention is that without such internal reference reaction, accurate PMR can be obtained for each independent sample regardless of the total amount of starting DNA template. In addition, we compared the reproducibility of these two detection methods in 10 clinical tumor samples (the specific steps of traditional MethyLight refer to Eads CA, Danenberg K...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com