Engineering bacterium for degrading polycyclic aromatic hydrocarbon, engineering reforming method and application thereof
A technology of polycyclic aromatic hydrocarbons and engineering bacteria, which is applied in the fields of genetic engineering and biodegradation, can solve the problems of difficult conversion of Pseudomonas bacteria and low conversion rate of heat shock, and achieve good degradation ability and improve the effect of degradation ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Clone catA2 gene from Pseudomonas aeruginosa SA1
[0029] (1) Extract the whole genome and plasmid of Pseudomonas aeruginosa SA1;
[0030] (2) According to the degradation gene sequence encoding polycyclic aromatic hydrocarbon ring lyase, design specific primers to specifically primer catAF and catAR.
[0031] catAF: 5'-ATGACCGTGAAGATATCTCACAC-3'
[0032] catAR: 5'-TTAGATTTCTTGCAAGGCTCTCG-3'
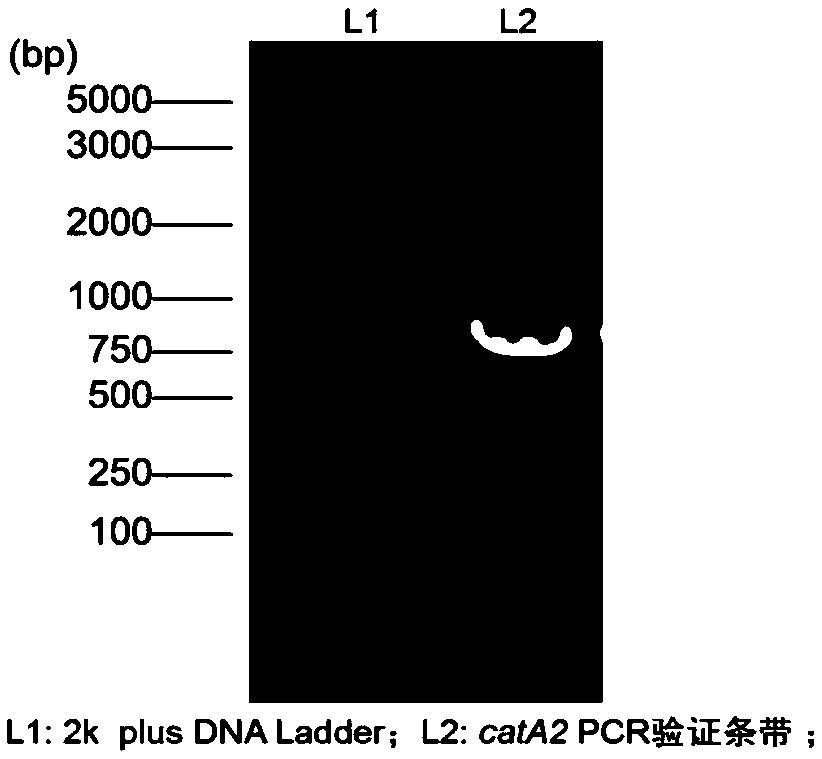
[0033] Using the genome and plasmid as a template, amplify a specific band of about 900bp, such as figure 2 As shown, the size corresponds to the size of the catA2 gene;
[0034] (3) Send the amplified product to Beijing Jinweizhi Biotechnology Co., Ltd. for sequencing, and the sequencing result is shown in SEQ ID No.1. Comparing the sequencing results with the catA2 gene encoding polycyclic aromatic hydrocarbon ring lyase, the results show that the two sequences are completely consistent, and it can be determined that the catA2 gene has been cloned from the bacteri...
Embodiment 2
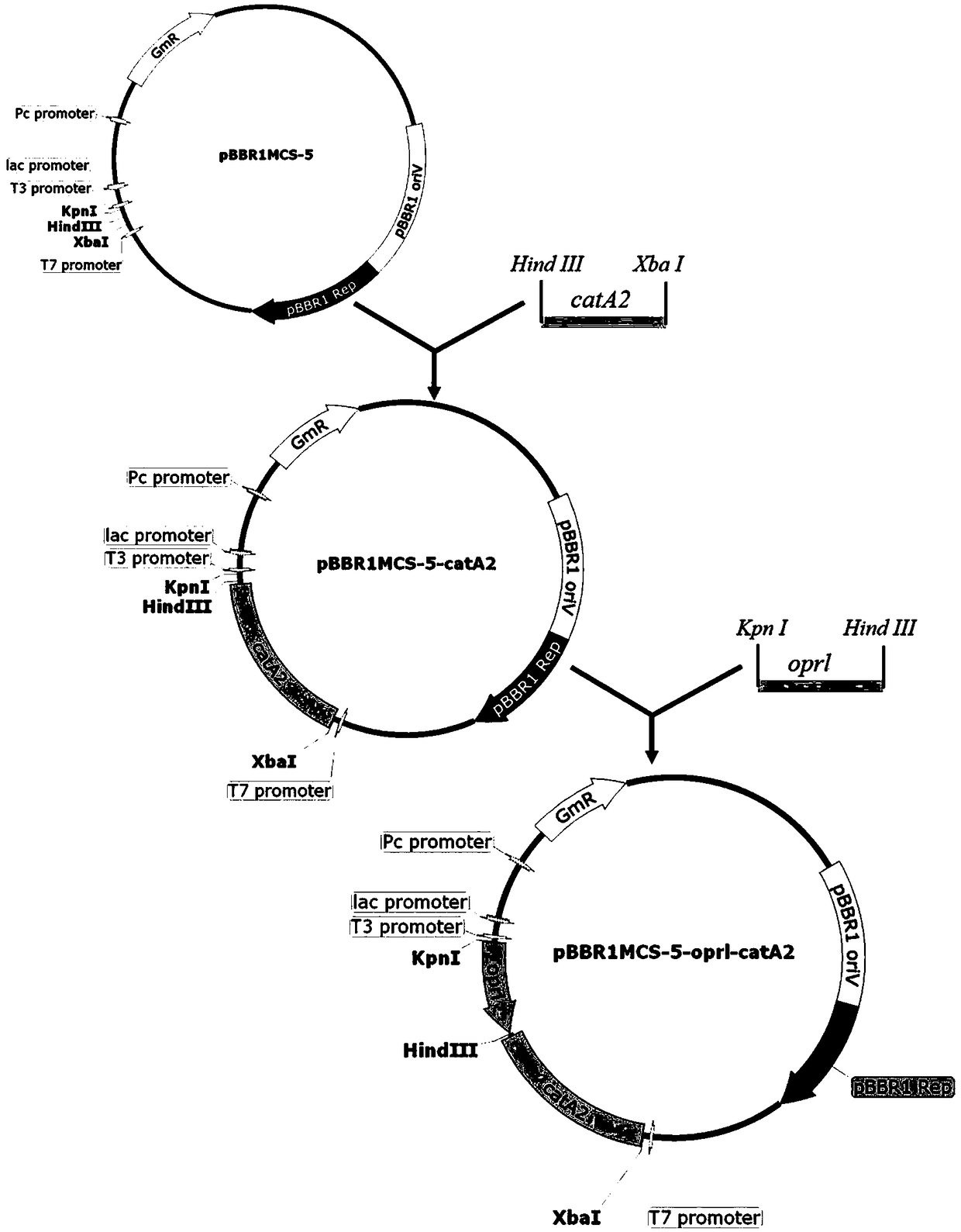
[0035] Example 2 Construction of recombinant expression vector pBBR1MCS-5-oprL-catA2
[0036] (1) For the degradation gene catA2, add HindⅢ and XbaⅠ restriction sites at both ends of the fragment by PCR method, and use FastDigest endonucleases HindⅢ and XbaⅠ to perform digestion. The reaction system is: 5 μL 10*FDbuffer, 2.5 μL HindⅢ, 2.5 μL XbaⅠ, 30 μL catA2 gene with restriction enzyme site and 10 μL ultrapure water. The reaction conditions are: 37°C, 1h. Similarly, the pBBR1MCS-5 plasmid was digested with FastDigest endonucleases HindⅢ and XbaI, purified and recovered using the PCR purification kit. The digested catA2 gene and pBBR1MCS-5 plasmid were ligated. The reaction system is: 1 μL 10*T4DNALigase Buffer, 1 μL T4DNA Ligase, 6 μL digested nucleotide fragments and 2 μL digested pRSFDuet plasmid. The reaction conditions are: 22°C, 10 min. After enzyme digestion and ligation, the competent cells E.coli DH5α were transformed, the positive clones were screened by colony ...
Embodiment 3
[0038] Embodiment 3 recombinant vector is transformed in Pseudomonas aeruginosa SA1
[0039] The detailed construction steps of recombinant expression vector pBBR1MCS-5-oprL-catA2 transformed into Pseudomonas aeruginosa chassis strain SA1 are as follows:
[0040] (1) Add 2.5 μL of the recombinant expression vector pBBR1MCS-5-oprL-catA2 to 50 μL of Pseudomonas aeruginosa SA1 competent cells, and gently pipette to mix;
[0041] (2) Put the mixture into a 2mm electric cup and shock at 2.5KV for 5-6ms. Then quickly add 1mL anti-resistant LB medium, recover at 37°C, 200rpm for 2h, spread gentamicin-resistant LB solid medium plate, and culture overnight at 37°C;
[0042] (3) The positive transformants verified by colony PCR were selected and cultured overnight in 5 mL LB medium to obtain engineering Pseudomonas aeruginosa PH2, and the bacterial strain was preserved.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com