Aptamer capable of being specifically combined with nodularin-R and application of the aptamer
An aptamer and specific technology, which is applied in the field of aptamers that specifically bind to nodularin-R, achieves the effects of high repeatability, simplified construction of technical routes, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1. The design of random ssDNA library and its primers
[0038] 1. Design of ssDNA library
[0039] The NOD-R aptamer library consists of 80 bases, both ends are fixed regions containing 20 bases, and the middle is a random region containing 40 bases: 5'-AAGGAGCAGCGTGGAGGATA-N40-TTAGGGTGTGTCGTCGTGGT-3' (N is any one of the four deoxyribonucleotide bases of A, T, G, and C, and 40 represents the number of random bases).
[0040] 2. Primer Design
[0041] Upstream primer: 5'-AAGGAGCAGCGTGGAGGATA-3' (SEQ ID NO.13)
[0042] Downstream primer A: 5'-ACCACGACGACACACCCTAA-3' (SEQ ID NO.14)
[0043] Downstream primer B: 5'-poly(dA20)-Spacer18-ACCACGACGACACACCCTAA-3' (SEQ ID NO.15).
[0044] Among them, the downstream primer A is mainly used for the amplification of ssDNA in the screening, and the downstream primer B is mainly used for cloning and sequencing.
Embodiment 2
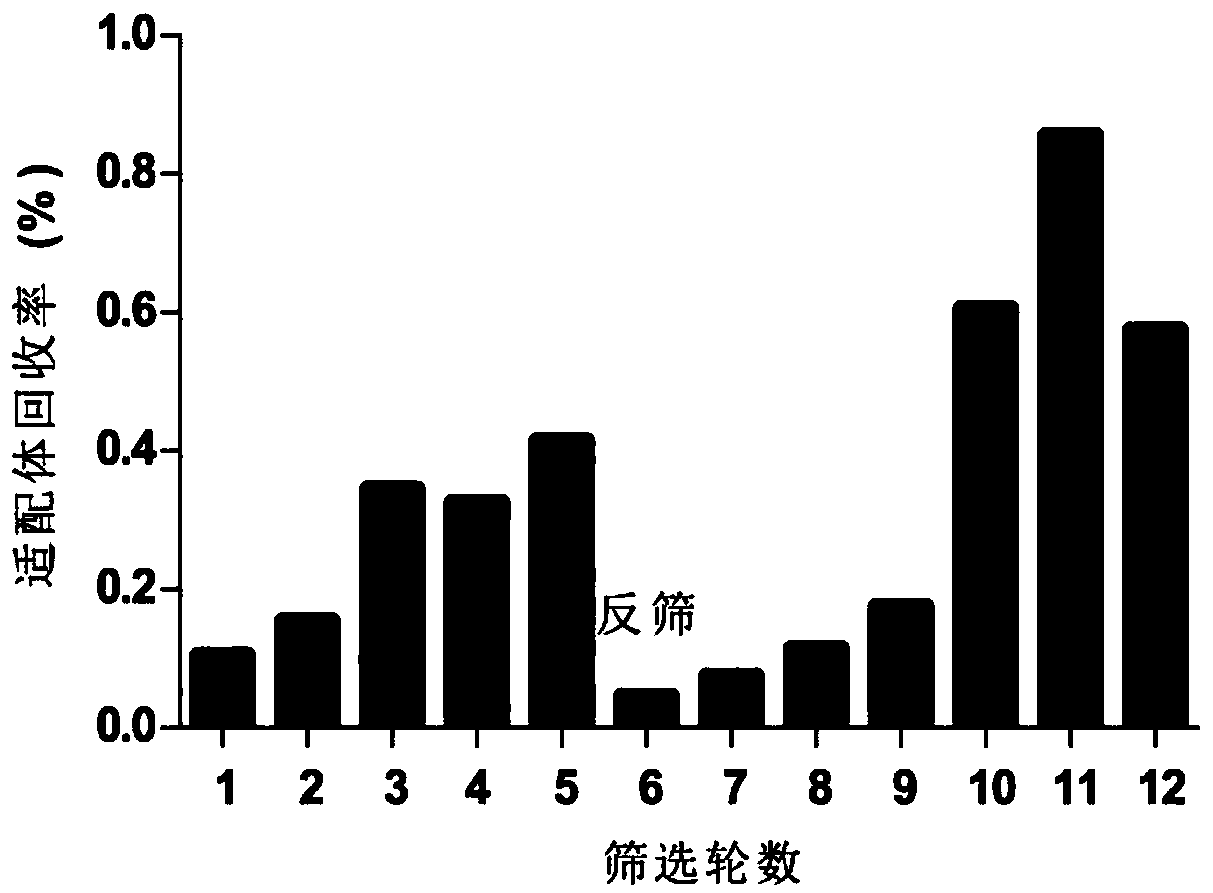
[0045] Example 2. Screening of NOD-R aptamers
[0046] According to the characteristics of NOD-R molecules, the classical aptamer screening method—magnetic bead method was used for screening.
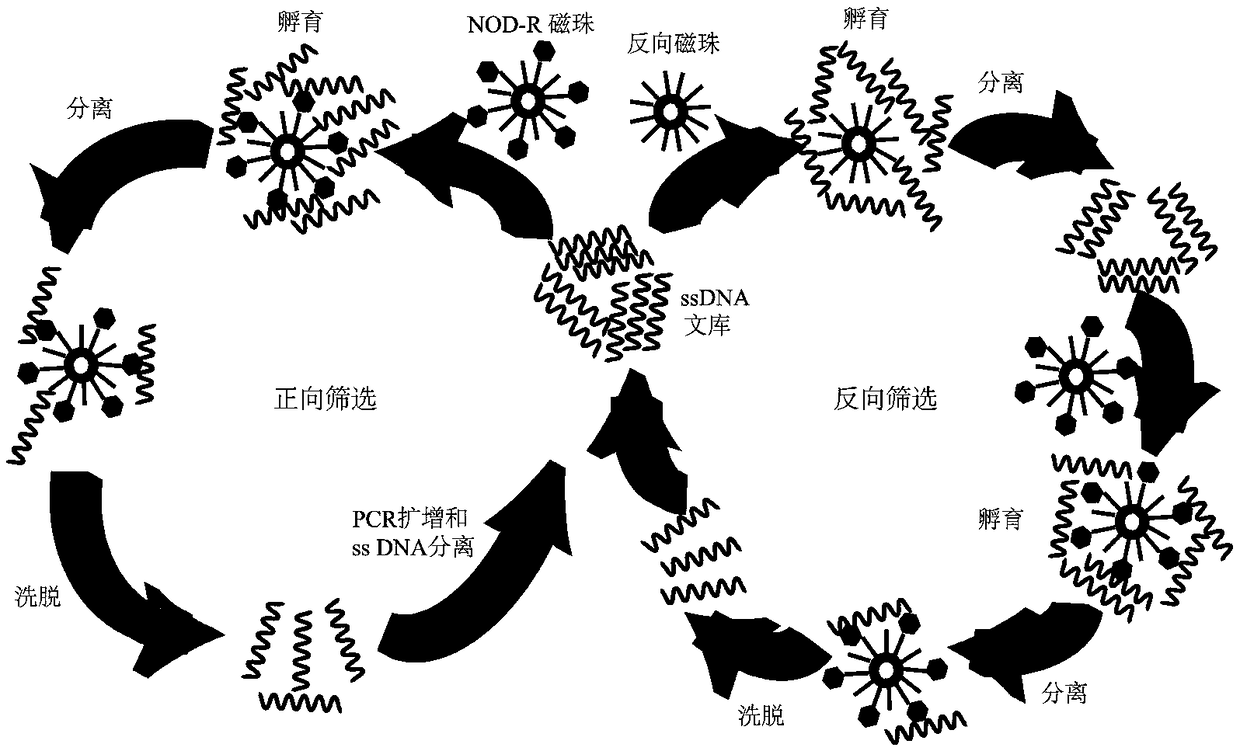
[0047] Such as figure 1 As shown, the screening process mainly includes four steps, namely incubation, separation, elution and amplification. Table 1 shows the specific screening scheme, and the specific process is as follows:
[0048] (1) Incubation: Take 3 nmol ss DNA library (the second to fifth round library volume is 200 pmol, and the sixth to twelfth round library volume is 120 pmol) for denaturation treatment, that is, first heat in a 95°C water bath. Bath for 10 minutes; then, quench in an ice bath for 5 minutes; finally, place at room temperature for 5 minutes; at the same time, wash the NOD-R magnetic beads several times with screening buffer, and add 20 μL to the ssDNA library that has been treated above; add a certain amount of screening buffer to a total volume of 600 μL ...
Embodiment 3
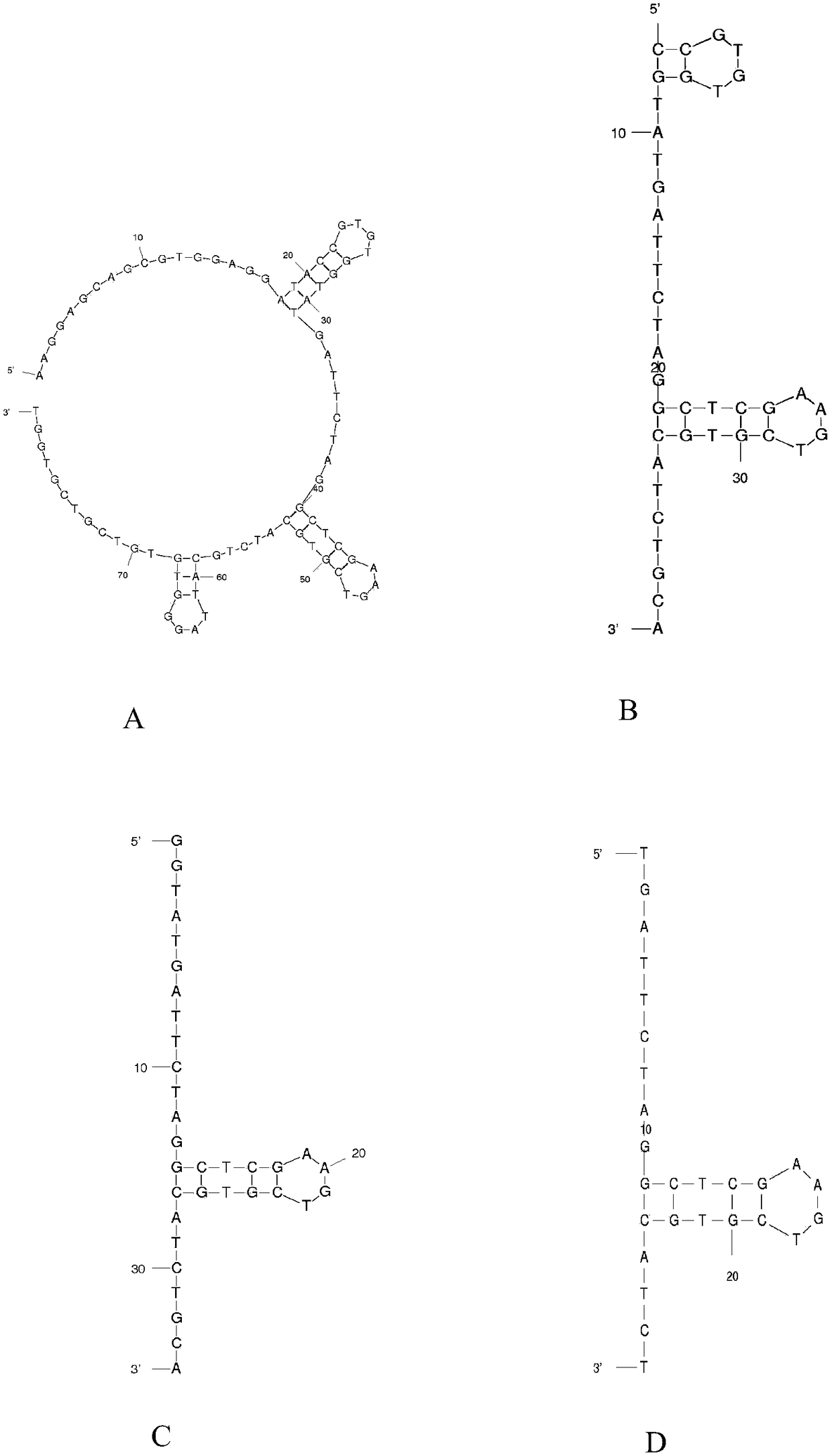
[0061] Example 3. Optimization of aptamer N13
[0062] Among the selected 10 aptamers, the affinity constant (K D ) has the smallest value, which is 0.138μM. In order to obtain the core structure of aptamer N13, the aptamer was truncated and optimized.
[0063] image 3 It shows the predicted secondary structure of the truncated aptamer N13 by the online tool the mfold web server.
[0064] When the fixed regions at both ends of the aptamer N13 are removed (N13-T), the aptamer can still bind to NOD-R, and K D The value is the same as before truncation. Therefore, it can be inferred that the primer-binding regions at both ends of the aptamer N13 are not involved in the combination with NOD-R.
[0065] According to the prediction results of the secondary structure of aptamer N13-T by the online tool the mfoldweb server, it was further truncated, and aptamer N13-T-O1, aptamer N13-T-O2, aptamer N13-T-O2, Aptamer N13-T-O3 and Aptamer N13-T-O4. Among the four aptamers, except ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com