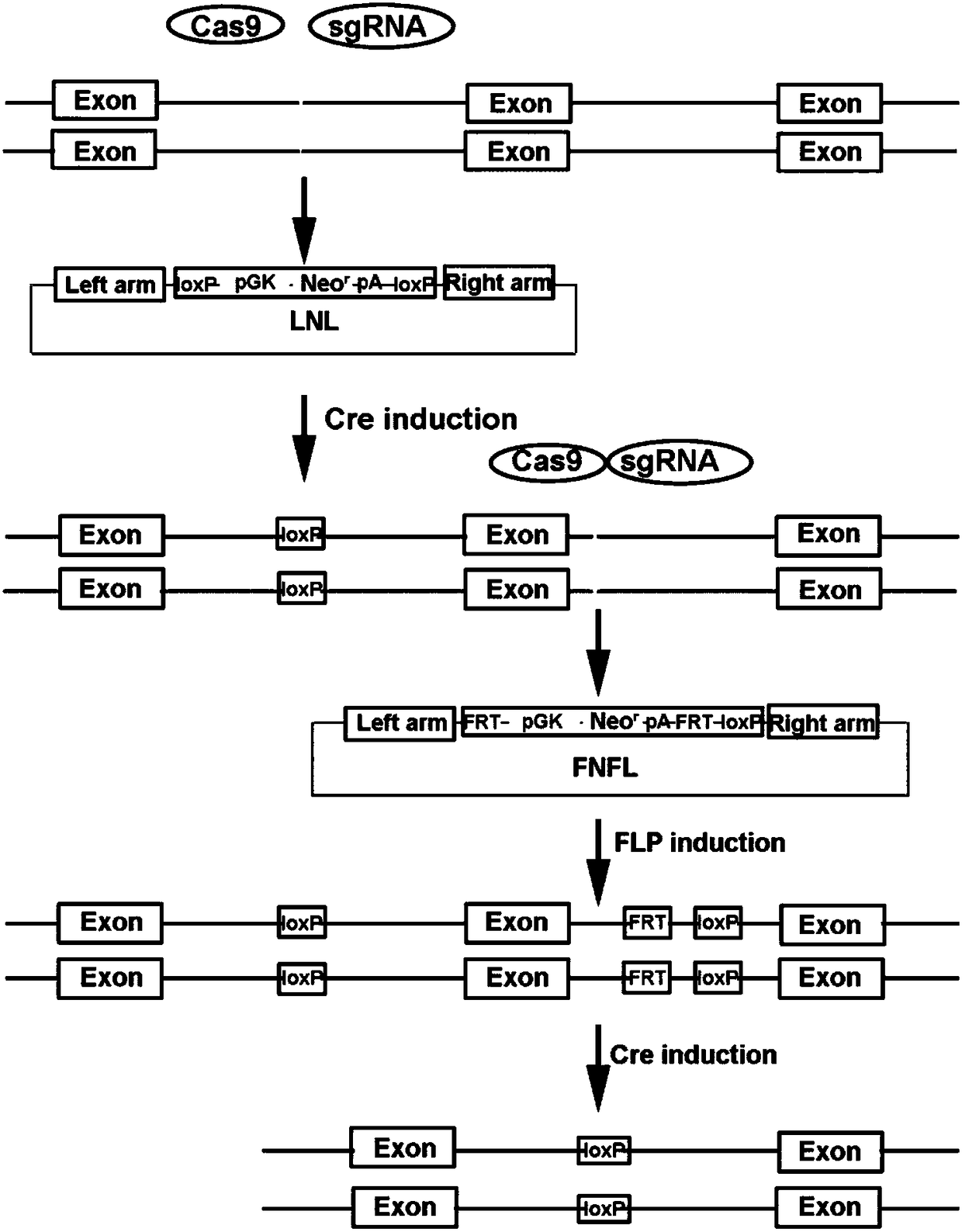
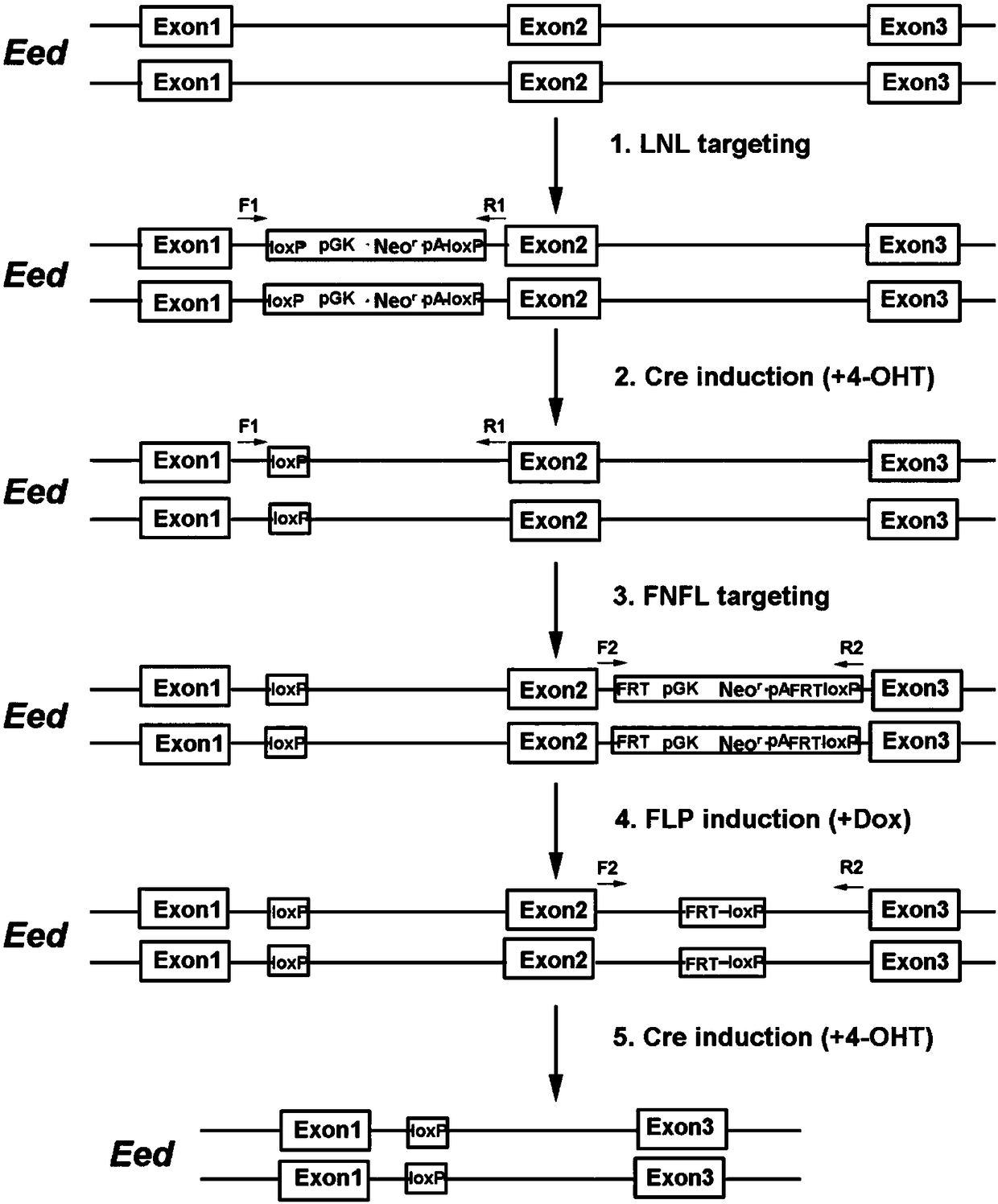
Conditional gene knockout method constructed by using CRISPR/Cas9 system
A conditional and genetic technology, applied in the field of gene knockout, can solve the problems of homology arm length, affect the efficiency of conditional gene knockout, and low recombination efficiency, so as to improve the success rate, save time and labor costs, and improve efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] The present embodiment provides a method for selectively knocking out the Eed gene (gene of interest) in mouse embryonic stem cells (cell of interest), specifically comprising the following steps:
[0050] (1) Construction of the left targeting plasmid of the Eed gene: select the LNL sequence to be inserted at the appropriate position upstream of exon 2 of the Eed gene, select the left and right homology arms ranging from 500bp to 1kb for the upstream and downstream of the site, and use high-fidelity Phusion DNA polymerase (M0530S, NEB) amplifies the left and right homology arms of the LNL sequence to be inserted, and then purifies it with AxyPrep PCR purification kit. The left arm uses KpnⅠ and EcoRI restriction endonucleases to digest the PCR product, and the right arm uses BamHI and SacⅡ digested, inserted into the plasmid containing the LNL sequence, and constructed the targeting plasmid on the left side of the target gene.
[0051] (2) Use the website http: / / crispr...
Embodiment 2
[0060] This embodiment provides a method for conditionally knocking out the SRCAP gene (gene of interest) in mouse embryonic stem cells (cell of interest), specifically comprising the following steps:
[0061] (1) Construction of the LNL targeting plasmid on the left side of the SRCAP gene knockout region: select exons 5-8 of the SRCAP gene as the target knockout region, and use a position upstream of exon 5 as the LNL insertion site. First, use the mouse genome as a template, use Pusion high-fidelity DNA polymerase to amplify the left and right homology arms of 500bp to 1kb in size upstream and downstream of the insertion site, and then use AxyPrep PCR purification kit to purify; then restrict with KpnⅠ and EcoRI The left-arm PCR purified product was digested with a neutral endonuclease, and the right arm was digested with BamHI and SacII, and then the two homologous arms were sequentially connected to the LNL vector to obtain the LNL plasmid targeting the left side of the tar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com