Recombinant escherichia coli capable of efficiently producing glucosamine and application thereof
A technology of glucosamine and glucose, applied in the field of bioengineering, can solve the problems of high industrialization cost, low yield, high glucose, etc., and achieve the effects of low cost, increased yield, and high-efficiency expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
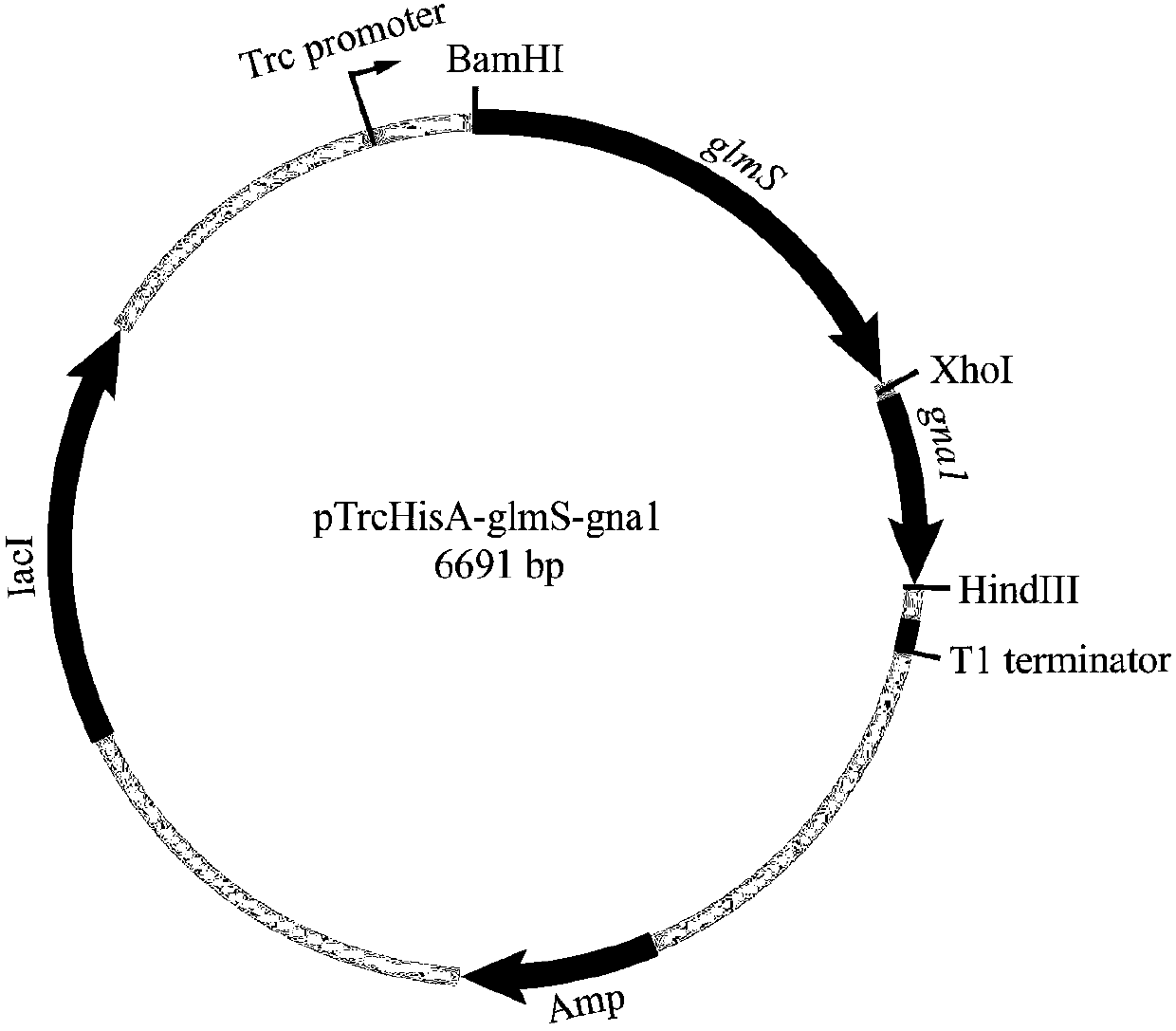
[0040] Embodiment 1: Construction of expression vector pTrcHisA-glmS
[0041] The glucosamine synthase gene glmS used in the present invention is derived from E. coli MG1655, which is a commercial strain. Using pTrcHisA as the expression vector. pTrcHisA contains a trc promoter, which can enable the normal expression of the target gene in E. coli MG1655, while the commonly used pET series of plasmids are composed of T7 promoters, which cannot express the target gene in E. coli MG1655, so the pTrcHisA plasmid was selected.
[0042] E.coli MG1655 was inoculated in 25mL LB liquid medium, cultured at 37°C, 200rpm for 12h, the bacteria were collected, and the genomic DNA of the E. coli strain was extracted using a bacterial genome extraction kit.
[0043] According to the published genome information sequence, respectively design sequences such as SEQ ID NO.6 / SEQ ID NO.7 primer pair glmS-S / glmS-A, using the extracted genomic DNA as a template, using standard PCR amplification syst...
Embodiment 2
[0047] Example 2: Construction of expression vector pTrcHisA-glmS-gna1
[0048] The glucosamine acetylase gene gna1 used in the present invention is derived from S.cerevisiae BY4741, and S.cerevisiae BY4741 is inoculated in 25mLYeast Extract Peptone Dextrose Medium (YPD) liquid medium, cultivated at 30°C and 200rpm for 12h, and the bacteria are collected and used The Fungal Genome Extraction Kit extracts genomic DNA from yeast strains.
[0049] According to the published genome information sequence, respectively design sequences such as SEQ ID NO.8 / SEQ ID NO.9 primer pair gna1-S / gna1-A, using the extracted genomic DNA as a template, using standard PCR amplification system and program , to amplify and obtain the gna1 gene.
[0050] gna1-S (SEQ ID NO.8):ACTCGAGATGAGCTTACCCGATGGATTTTATATAAG
[0051] gna1-A (SEQ ID NO.9):AAGCTTCTATTTTCTAATTTGCATTTCCACGC
[0052] The gna1 obtained by PCR amplification was recovered by agarose gel electrophoresis, and the recovered product and th...
Embodiment 3
[0053] Example 3: Construction of the gene nagE genome editing fragment that weakens the acetylglucosamine phosphate transporter
[0054] In order to realize the translation of the acetylglucosamine transporter gene nagE, the initiation codon ATG was optimized by replacing it with TTG, and the genome editing fragment was constructed with the help of Red homologous recombination technology. The gene editing fragments include upstream and downstream homology arm regions, and resistance screening cassettes. Using the E.coli MG1655 genome as a template, design a sequence such as SEQ ID NO.10 / SEQ ID NO.11 primer pair nagE-S1 / nagE-A1 to amplify the nagE gene, and use the plasmid pKD4 as a template to design a sequence such as SEQ ID NO. 12 / SEQID NO.13 primer pair nagE-S2 / nagE-A2 amplification resistance screening gene Kan, and then the above PCR products were recovered by agarose nucleic acid gel electrophoresis, and the recovered nagE gene and Kan were used as templates, NagE-S1 / n...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com