A Nucleic Acid Analysis Method Based on Cascade Hybridization Chain Reaction
A hybrid chain reaction, cascade technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as unresearched
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
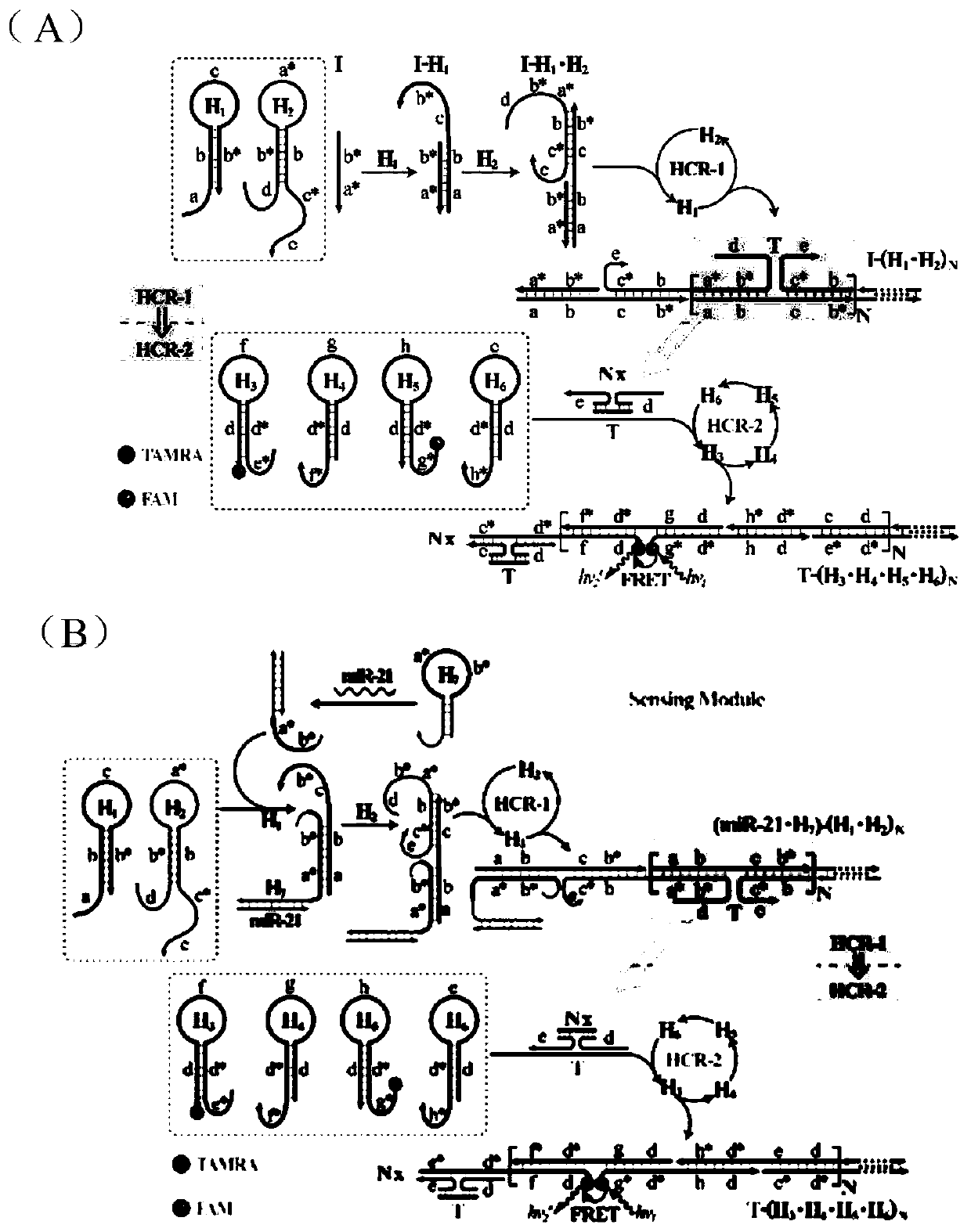
[0042] Design of hairpin probes: Use NUPACK software to design relevant hairpin probes, and entrust Sangon Bioengineering (Shanghai) Co., Ltd. to synthesize relevant nucleic acid sequences. When there is no target, ensure that the stem end of each hairpin has enough complementary base pairing to maintain its own stability and keep the fluorescence unchanged, but when there is a target, it can trigger a cascade hybridization chain reaction and form a branch. Shaped DNA nanostructures undergo fluorescence resonance energy transfer to achieve target detection. All hairpin probe dry powders were first dissolved in phosphate buffer, and their absorbance was measured with a UV spectrophotometer to calculate the exact concentration. 2 ) Prepare all the hairpin probes at 4 μM, in PCR at 95° C. for 5 minutes, and at 25° C. for 2 hours to form stable hairpins. All reactions were carried out in HEPES buffer.
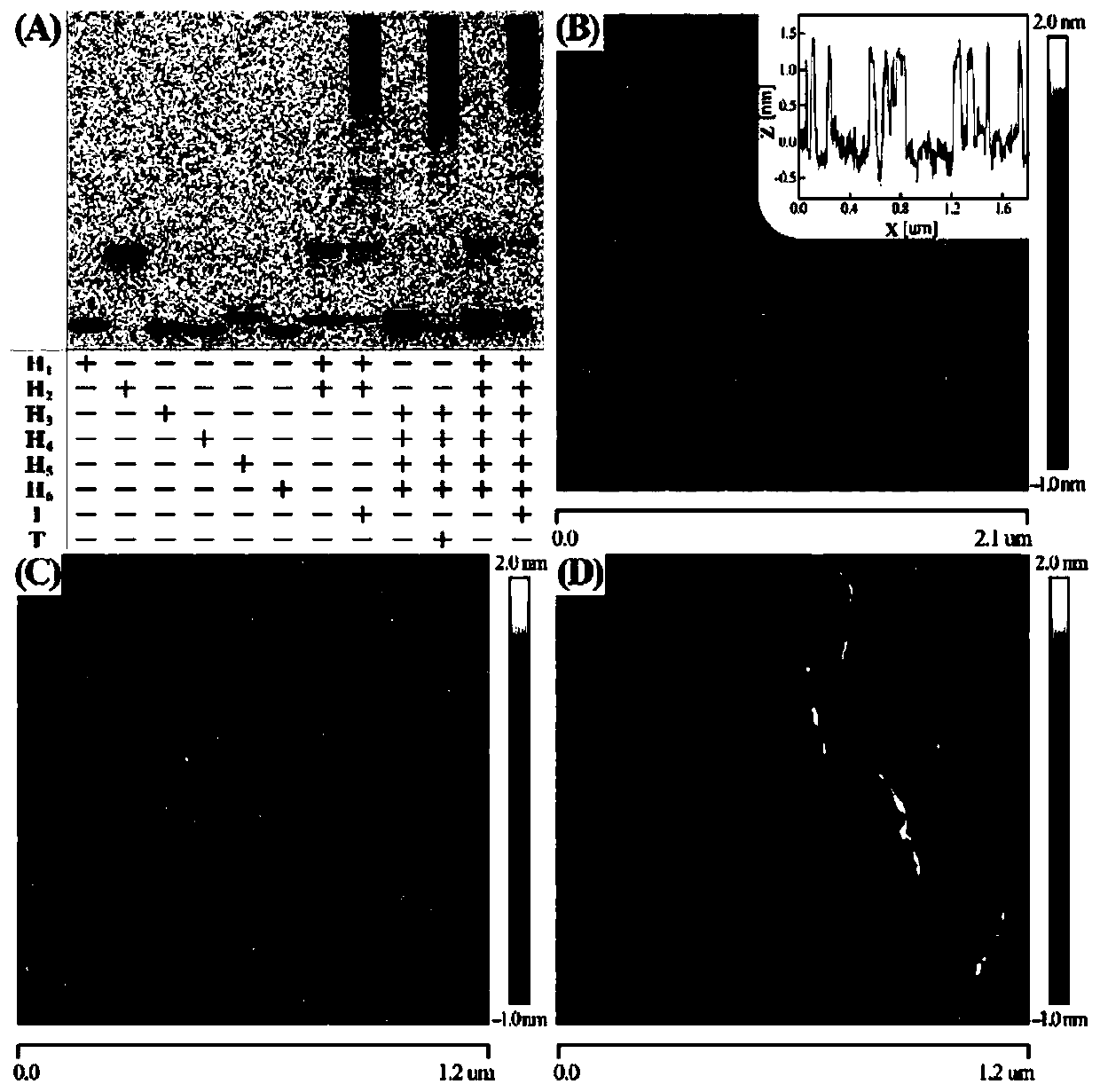
[0043] figure 1 A is the schematic diagram of cascade hybridization chain r...
Embodiment 2
[0046] [Example 2] DNA detection based on cascade hybridization chain reaction
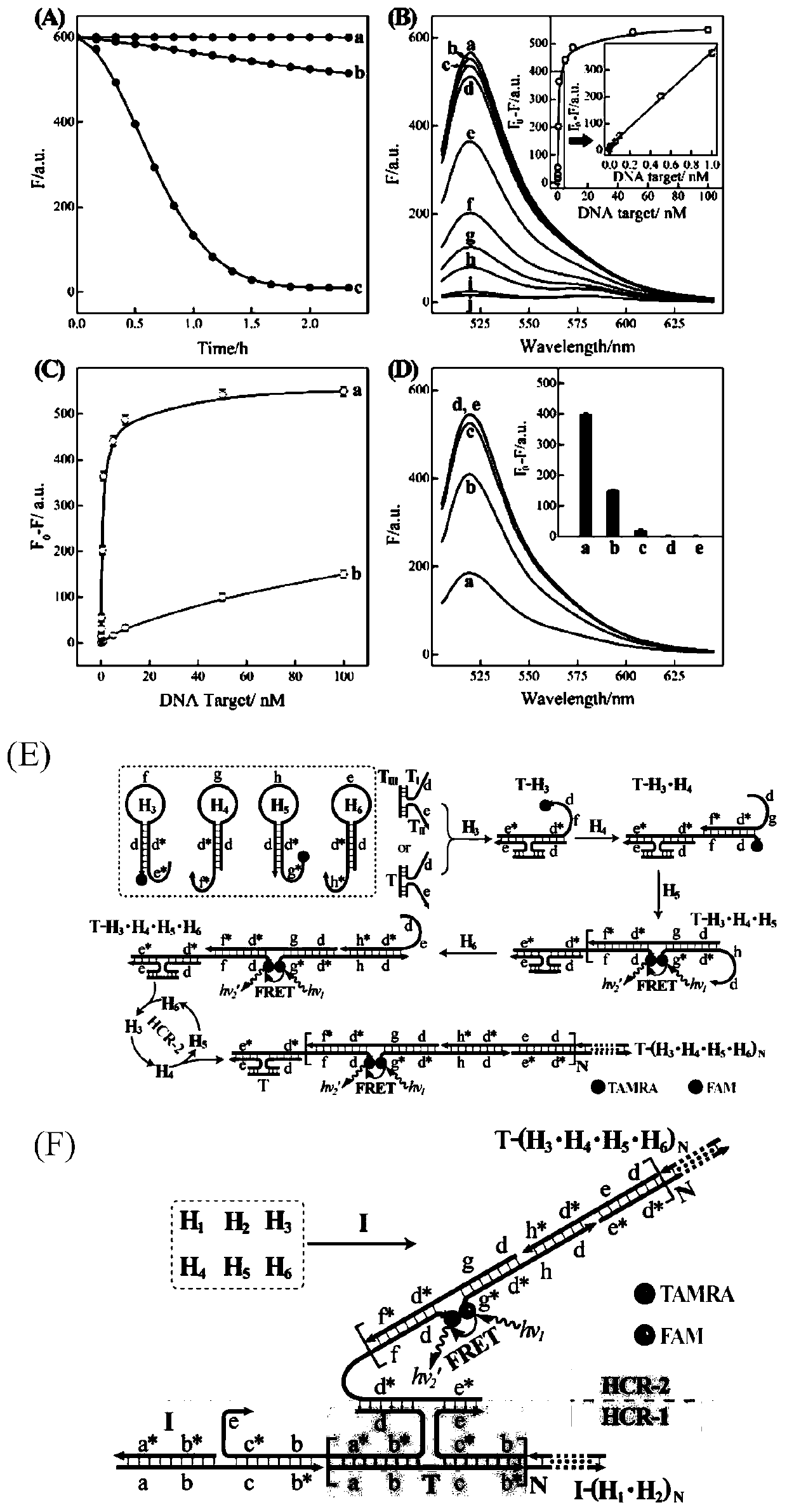
[0047] In hydroxyethylpiperazine ethyl sulfonate buffer (concentration is 10mM, pH 7.2, containing 1M NaCl and 50mM MgCl 2 ), set all hairpin probes (H 1 、H 2 、H 3 、H 4 、H 5 and H 6 Both are 200nM) with different concentrations of target DNA (0, 1×10 -11 , 5×10 -11 , 1×10 -10 , 5×10 -10 , 1×10 -9 , 5×10 -9 , 1×10 -8 , 5×10 -8 , 1×10 -7 M) Mixing, incubating at room temperature for 2h, using a fluorescence spectrometer to measure the fluorescence intensity of the system (excitation voltage 600V, excitation slit 5nm, emission slit 10nm, excitation wavelength 490nm, wavelength scanning range 505-650nm).
[0048] Depend on figure 2 (B) It can be seen that when no target DNA is added to the C-HCR system, each hairpin probe can maintain its own stability, and the fluorescence of the system will not change ( figure 2 Curve a) in (B), when different concentrations of target DNA are added...
Embodiment 3
[0051] [Example 3] In vitro detection of miRNA-21 based on cascade hybridization chain reaction
[0052] In hydroxyethylpiperazine ethyl sulfonate buffer (concentration is 10mM, pH 7.2, containing 1M NaCl and 50mM MgCl 2 ), set all hairpin probes (H 1 、H 2 、H 3 、H 4 、H 5 、H 6 Both are 200nM, H 7 50nM) with different concentrations of miRNA-21 (0, 1×10 -11 , 5×10 -11 , 1×10 -10 , 5×10 -10 , 1×10 -9 , 5×10 -9 , 1×10 -8 , 5×10 -8 , 1×10 -7 M) Mixing, incubating at room temperature for 2h, using a fluorescence spectrometer to measure the fluorescence intensity of the system (excitation voltage 600V, excitation slit 5nm, emission slit 10nm, excitation wavelength 490nm, wavelength scanning range 505-650nm).
[0053] figure 1 B is the schematic diagram of cascade hybridization chain reaction for miRNA-21 detection. Depend on Figure 4 (A) It can be seen that when miRNA-21 is not added to the C-HCR system, each hairpin probe can maintain its own stability, and the fl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com