LAMP primer combination for detecting 8 environmental pathogens of dairy cow mastitis and application thereof
A primer combination and primer set technology, which is applied in the direction of microorganism-based methods, microorganisms, and microorganism determination/inspection, can solve the problems of long PCR detection time, heavy workload, and low sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0235] Embodiment 1, the preparation of kit
[0236] The kit consists of eight LAMP primer sets, each for the detection of one environmental pathogen of dairy mastitis.
[0237] The primer set used to detect E. coli is as follows (5'→3'):
[0238] Outer primer F3 (SEQ ID NO: 1): TGTGGAATTGATCAGCGTT;
[0239] Outer primer B3 (SEQ ID NO: 2): TGATTATTGACCCACACTTTG;
[0240] Internal primer FIP (SEQ ID NO: 3): TCTGCATCGGCGAACTGATCGGGTGGGAAAGCGCGTTAC;
[0241] Internal primer BIP (SEQ ID NO: 4): TCTGGTATCAGCGCGAAGTCTAATGAGTGACCGCATCGA;
[0242] Loop primer LF (SEQ ID NO: 5): GCAATTGCCCGGCTTTCTT;
[0243] Loop primer LB (SEQ ID NO: 6): GCAGGCCAGCGTATCGT.
[0244] The primer set used to detect Klebsiella pneumoniae is as follows (5'→3'):
[0245] Outer primer F3 (SEQ ID NO: 7): ATACAAAAACACCAGTGTAGG;
[0246] Outer primer B3 (SEQ ID NO: 8): GCCGCCAGTTTGTTTCAG;
[0247] Internal primer FIP (SEQ ID NO: 9): CGTTGAGATTTGCGAAGTACCAAGAATCAAATATGCTGCAAATGTG;
[0248] Internal prime...
Embodiment 2
[0294] Embodiment 2, specificity
[0295] Test sample 1: Escherichia coli ( 8739 TM ).
[0296] Test sample 2: Klebsiella pneumoniae ( 13883 TM ).
[0297] Test sample 3: Pseudomonas aeruginosa (CVCC 3359).
[0298] Test sample 4: Streptococcus dysgalactiae ( 12388 TM ).
[0299] Test sample 5: Streptococcus uberis ( 700407 TM ).
[0300] Test sample 6: Salmonella typhimurium ( 14028 TM ).
[0301] Test sample 7: Proteus mirabilis (CVCC 1969).
[0302] Test sample 8: Candida albicans (CGMCC 2.2086).
[0303] Each sample to be tested carries out the following steps respectively:
[0304] 1. Extract the genomic DNA of the sample to be tested.
[0305] 2. Using the genomic DNA extracted in step 1 as a template, each primer set prepared in Example 1 was used to perform loop-mediated isothermal amplification.
[0306] Reaction system (10 μL): 7.0 μL reaction solution (product of Boao Bio Group Co., Ltd., catalog number CP.440020), 1 μL primer mixture, 1 μL te...
Embodiment 3
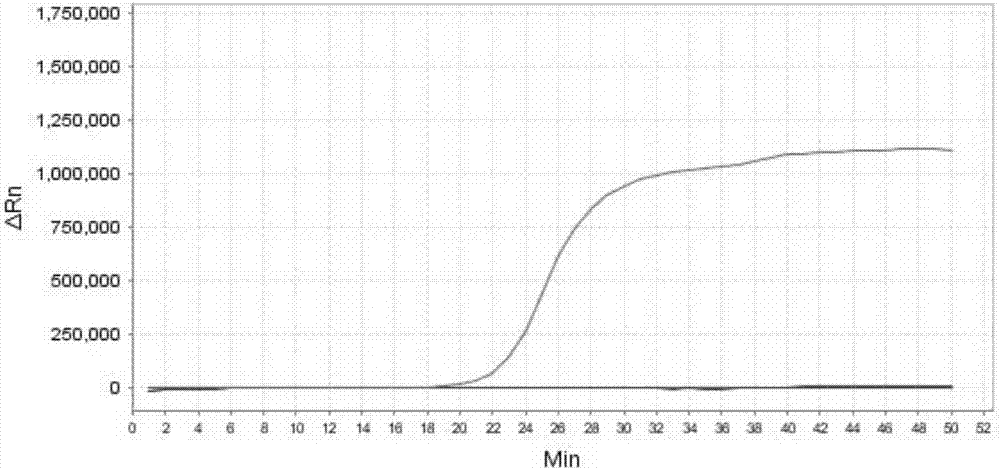
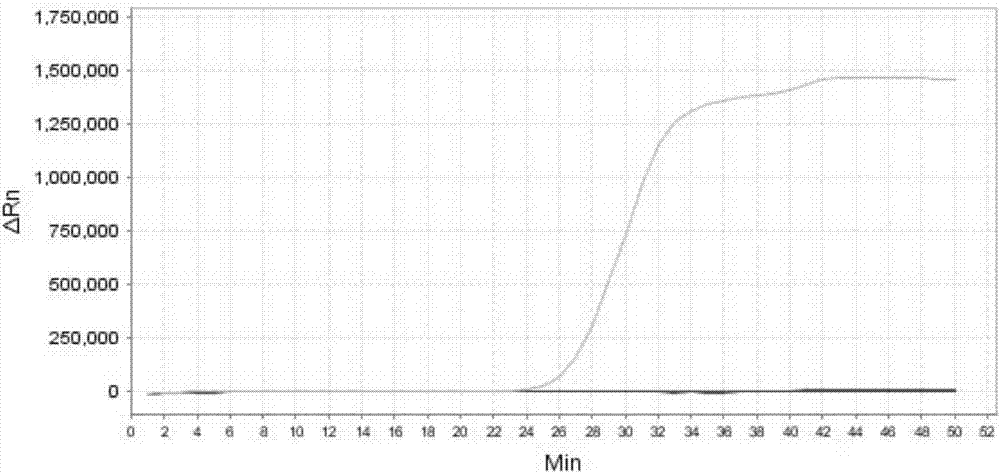
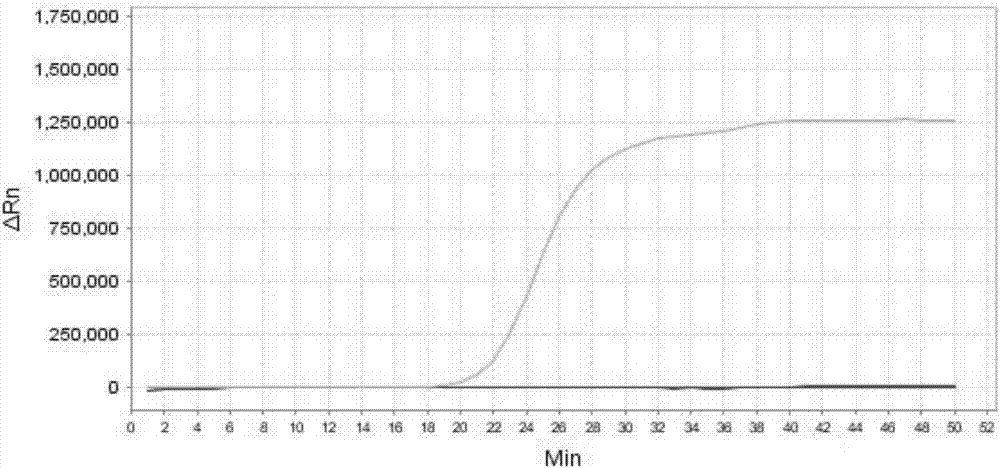
[0318] Embodiment 3, sensitivity result
[0319] Test sample 1: Escherichia coli ( 8739 TM ).
[0320] Test sample 2: Klebsiella pneumoniae ( 13883 TM ).
[0321] Test sample 3: Pseudomonas aeruginosa (CVCC 3359).
[0322] Test sample 4: Streptococcus dysgalactiae ( 12388 TM ).
[0323] Test sample 5: Streptococcus uberis ( 700407 TM ).
[0324] Test sample 6: Salmonella typhimurium ( 14028 TM ).
[0325] Test sample 7: Proteus mirabilis (CVCC 1969).
[0326] Test sample 8: Candida albicans (CGMCC 2.2086).
[0327] 1. Extract the genomic DNA of the sample to be tested, and perform gradient dilution with sterile water to obtain each dilution.
[0328] 2. Using the dilution obtained in step 1 as a template, the primer sets prepared in Example 1 were used to perform loop-mediated isothermal amplification.
[0329] When the sample to be tested is the sample to be tested 1, the loop-mediated isothermal amplification is performed using primer set I. When the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com