A detection method and application of human y chromosome label site sy1291
A technology of Y chromosome and detection method, applied in the field of detection of human Y chromosome tag site sY1291, can solve problems such as deletion of AZFc region, and achieve the effects of expanding detection range, rapid detection, and high deletion consistency rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
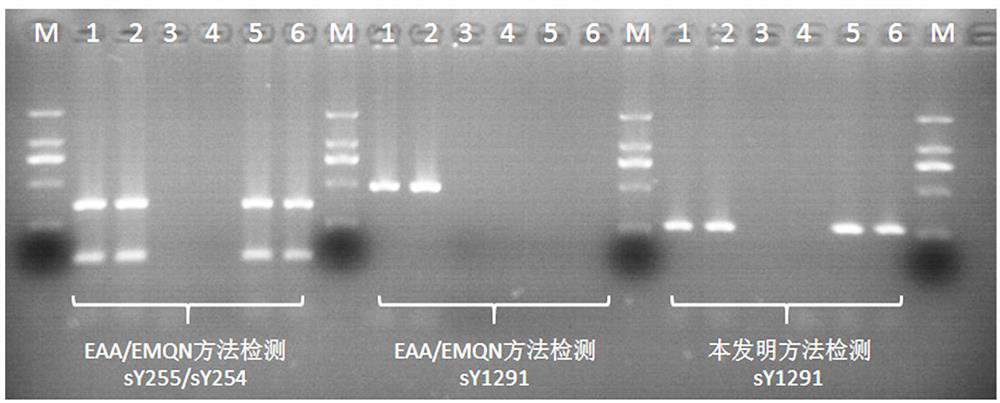
[0031] Example 1: The present invention combines PCR product agarose electrophoresis for sY1291 site detection
[0032] Using the PCR primers shown in SEQ ID No.1 and SEQ ID No.2 (wherein SEQ ID No.2 does not need to be labeled with fluorescein), the male genomic DNA was carried out according to the PCR reaction system shown in Table 2 and the PCR reaction conditions shown in Table 3 Amplification, PCR amplification products were electrophoresed with 2.0% agarose.
[0033] A total of 6 male samples from No. 1 to No. 6 were divided into two types according to the detection results and phenotype of the classic deletion sites in the AZFc region (sY255 and sY254). Samples No. 3 and No. 4 were male infertility samples with classic deletion sites in the AZFc region , samples No. 1, No. 2, No. 5 and No. 6 are normal male samples with normal semen parameters. The detection method of the classic deletion sites (sY255 and sY254) in the AZFc region adopts the detection method recommende...
Embodiment 2
[0039] Example 2: The present invention combines PCR product capillary electrophoresis fluorescent fragment analysis to detect sY1291 site
[0040] Using PCR primers shown in SEQ ID No.1 and SEQ ID No.2 (wherein SEQ ID No.2 is labeled with FAM fluorescein), according to the PCR reaction system shown in Table 4 and the PCR reaction conditions shown in Table 5, to 1169 cases Male DNA samples were amplified by PCR, and the PCR products were subjected to capillary electrophoresis using a 3500Dx genetic analyzer.
[0041] The 1169 male samples included 232 male samples with normal semen parameters and 937 male infertile samples. All samples were identified by the kit approved by the China Food and Drug Administration for the detection of AZF microdeletions (human chromosome AZF region microdeletion nucleic acid kit (PCR-fluorescent probe method, National Machinery Note 20153400764) for the canonical mapping of the AZFc region. Points (sY255 and sY254) were detected, and the sY1291...
Embodiment 3
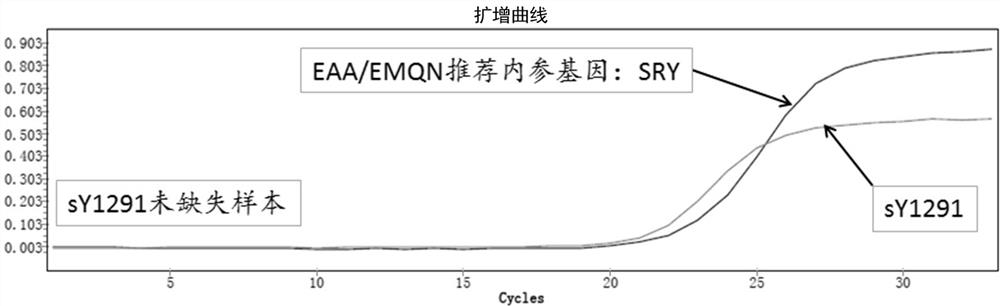
[0054] Example 3: The present invention combines the real-time fluorescent probe method to detect the sY1291 site
[0055] Using the PCR primers shown in SEQ ID No.1 and SEQ ID No.2 (SEQ ID No.2 does not label fluorescein), the fluorescent probe shown in SEQ ID NO.3, according to the PCR reaction system shown in Table 8 and Table 9 The PCR reaction conditions were shown for real-time fluorescence detection. The internal reference gene (sex-determining gene, SRY) recommended by EAA / EMQN for AZF microdeletion detection was detected in this system at the same time. When the fluorescent signal of the internal reference gene exists, the sample with sY1291 site amplification curve is the sample without sY1291 site deletion ( image 3 ), and those without sY1291 locus amplification curve are samples with sY1291 locus deletion ( Figure 4 ).
[0056] Table 8 PCR reaction system of the present invention combined with real-time fluorescence detection of sY1291 site
[0057] .
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com