Targeting sgRNA for pig CD163 gene editing, modification vector, and preparation method and application thereof
A gene modification and gene editing technology, applied in the field of genetic engineering, to ensure normal survival, reduce the work of genetically modified safety assessment, and have strong specific effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
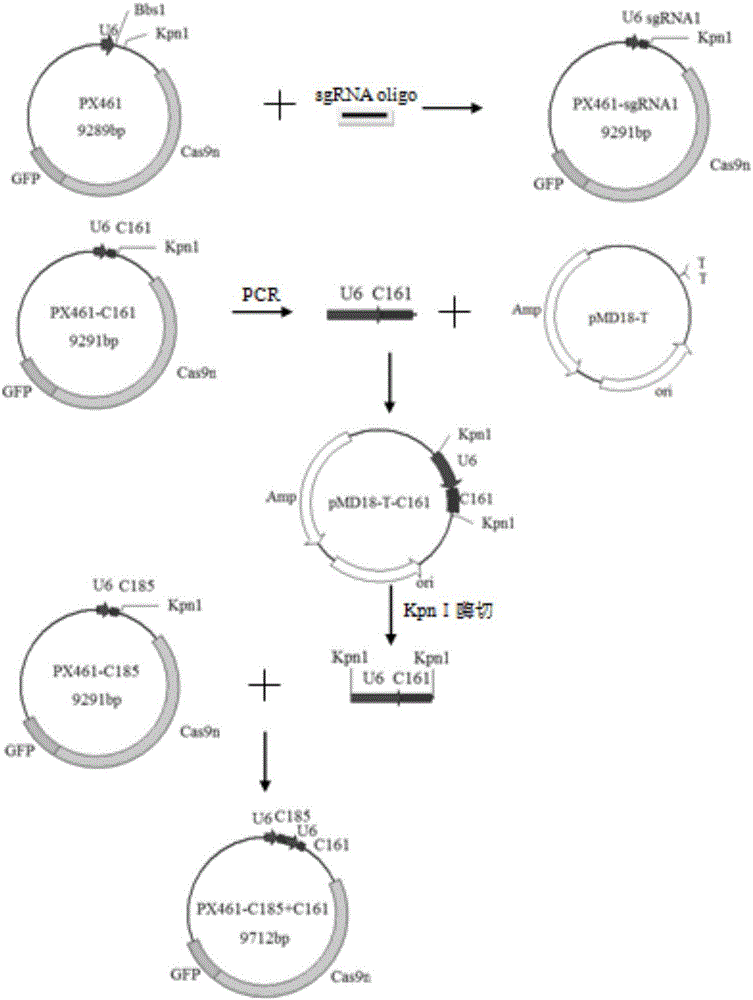
[0050] The present invention also provides a preparation method for the above-mentioned porcine CD163 gene modification carrier, comprising the following steps:
[0051] Step (a): Complementary pairing of the sense strand and the antisense strand of sgRNA-C161 to form a C161 double-stranded DNA molecule;
[0052] Step (b): Complementary pairing of the sense strand and the antisense strand of sgRNA-C185 to form a C185 double-stranded DNA molecule;
[0053] Step (c): connecting the C161 double-stranded DNA molecule to an expression vector to construct a vector expressing sgRNA-C161;
[0054] Step (d): connecting the C185 double-stranded DNA molecule to an expression vector to construct a vector expressing sgRNA-C185;
[0055] Step (e): Using the vector expressing sgRNA-C161 as a template, amplify the sgRNA-C161 sequence containing the U6 promoter by PCR, connect it to the cloning vector, and obtain the U6-sgRNA-C161 sequence after enzyme digestion;
[0056] Step (f): link the ...
Embodiment 1
[0072] The design of embodiment 1sgRNA and the construction of carrier
[0073] According to the seventh exon, the eighth exon and part of the upstream and downstream intron sequences (as shown in SEQ ID NO.5) and the mRNA sequence (NCBI NM_213976.1, as shown in SEQ ID NO.6) of the pig CD163 gene ), designing two sgRNAs on the seventh exon of the CD163 gene, sgRNA-C161 and sgRNA-C185, respectively adding cohesive linker sequences to both ends. Add the adapter sequence CACC to the 5' end of the F strand of each sgRNA sequence, and add the adapter sequence AAAC to the 5' end of the R strand of the reverse complementary sequence. If the first base at the 5' end of the sgRNA sequence is not G, then it should be First add a G to the 5' end of the F chain of the sgRNA sequence, and then add the linker sequence CACC. Correspondingly, add a C to the 3' end of the R chain of the reverse complementary sequence, so that it can be combined with the PX461 vector digested by BbsI The cohes...
Embodiment 2
[0101] Example 2 Transfection of pig somatic cells and screening of CD163 gene edited cell clones
[0102] Cell culture and transient transfection:
[0103] Pig fetal fibroblasts were inoculated in 6-well cell culture, and when they were cultivated to 50-70% confluence in DMEM medium containing 15% fetal bovine serum, according to the requirements of the instructions, Lipofectamine 2000 liposomes were provided in Example 1. The vector PX461-C185+C161 was transferred into the cells.
[0104] Flow cytometric sorting and monoclonal culture of transfected cells:
[0105] The transfected cells were cultured in a 37°C incubator for 48 hours, digested with 0.1% trypsin, sorted the positive cells expressing GFP by flow cytometry, and seeded the cells at a density of 50-100 cells / 100mm culture dish , cultured in DMEM medium containing 15% fetal bovine serum and 2.5ng / mL fibroblast growth factor (bFGF) for 9 to 12 days until obvious single-cell colonies appeared.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com