Method for directionally preparing alginate oligosaccharides by enzymatic hydrolysis
A technology of algin oligosaccharides and algin, which is applied in botany equipment and methods, biochemical equipment and methods, enzymes, etc., can solve the problems of low efficiency and inability to prepare algin oligosaccharides in a targeted manner, and achieves wide application prospects, Improve immunity, the effect of simple process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1: Preparation of algin oligosaccharides using alginase lyase before modification
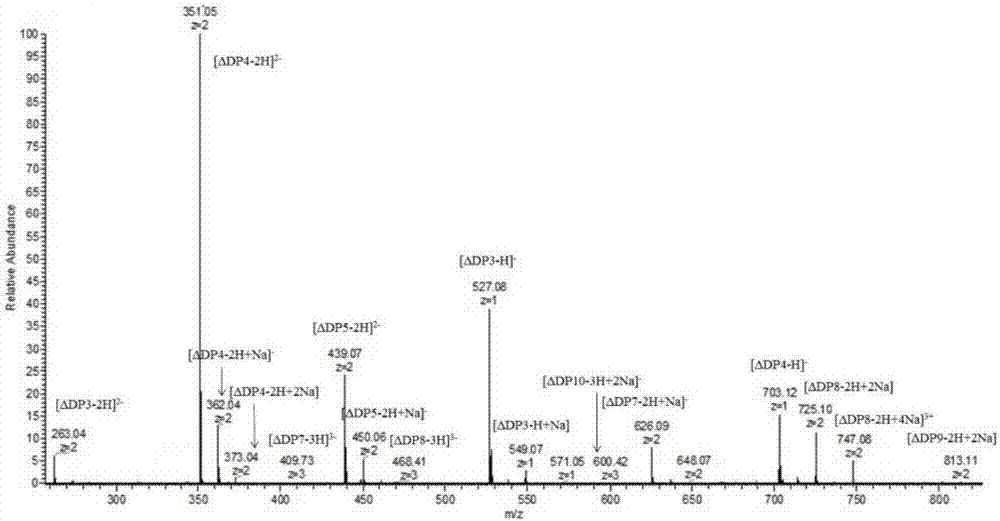
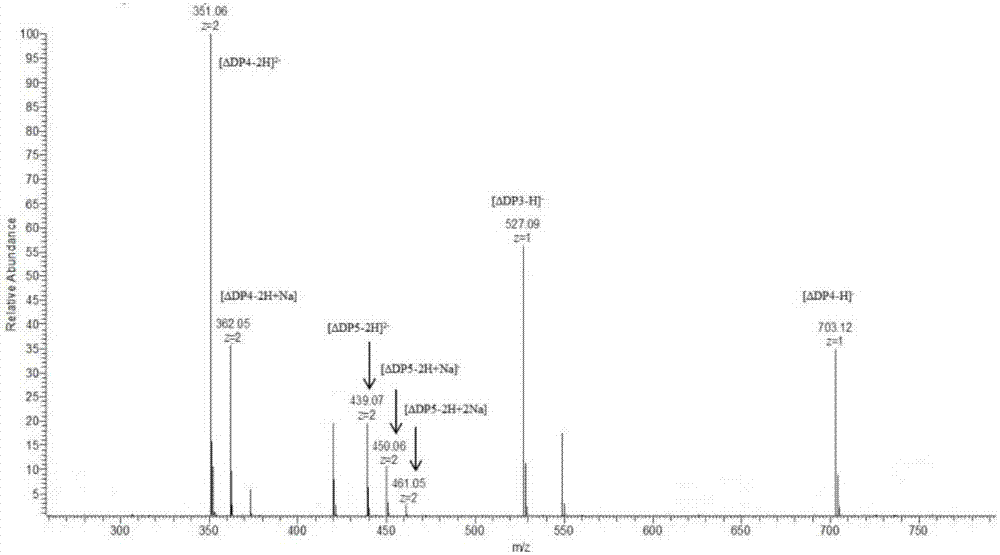
[0023] Dissolve alginate in water with pH=7 adjusted by NaOH, prepare 200 mL of alginate solution with a concentration of 5%, add 2 mL of alginate lyase (amino acid sequence such as: SEQ ID NO: 1, nucleotide sequence is SEQ ID NO: 2), stir and enzymolyze in a warm bath at 45°C for 2 hours, then add 2 mL of alginate lyase, continue to stir and enzymolyze in a warm bath at 45°C for 2 hours. The enzymolysis solution was centrifuged at 8000rpm for 10min to remove the residue, and the supernatant was the enzymolysis product. The enzymatic hydrolysis product was subjected to 4-fold alcohol precipitation to obtain the product, and the product was subjected to rotary evaporation and freeze-drying to obtain a crude alginate oligosaccharide with a yield of 43.9%. The product is detected by mass spectrometry, and the results show that the product includes a degree of polymerization of 3-10...
Embodiment 2
[0028] Example 2 Transformation of alginate lyase gene and construction of recombinant vector
[0029] In view of the low efficiency of enzymatic hydrolysis of alginate lyase, the wide distribution of enzymatic hydrolysis products, and the inability to directional produce target products, the applicant designed upstream primers based on the complete sequence of alginate lyase gene on the basis of further analysis of alginate lyase enzyme F1 and downstream primer R1 were amplified by PCR to obtain the complete sequence of the gene. The PCR conditions were: pre-denaturation at 94°C for 3 minutes, followed by 30 cycles of 94°C for 30s, 55°C for 30s, and 72°C for 2 minutes, and finally extension at 72°C for 5 minutes. Primers F1, R2 and F2, R1 were used to amplify the first 54 bases and the last 768 bases of the complete gene sequence respectively. The two PCR products were digested with AvrⅡ and ligated with T4 ligase to form a genetically modified sequence. The genetically modi...
Embodiment 3
[0031] Example 3 Production of Recombinant Alginate Lyase Using Escherichia coli Recombinant Strains
[0032] Pick the recombinant strain of Escherichia coli, inoculate it into 5 mL liquid LB medium containing 100 ug / mL ampicillin, and culture it with shaking at 170 r / min at 37°C for 12-16 hours. Inoculate 50mL of liquid LB medium containing 100ug / mL ampicillin according to 2% inoculum amount, shake culture at 37°C at 170r / min until OD600 is 0.5-0.8, add IPTG with a final concentration of 0.9mM, continue at 23°C at 150r / min Cultivate to 24h. Centrifuge at 1000 rpm for 10 min to obtain supernatant extracellular enzymes.
[0033] The enzyme activity was measured by the DNS method, and the results showed that the enzyme activity of the recombinant alginate lyase after genetic modification was 29.7 U / mL, which was 5.2 times that of the enzyme activity before genetic modification.
PUM
| Property | Measurement | Unit |
|---|---|---|
| degree of polymerization | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com