Agapanthus africanus cathepsin B, encoding gene thereof, probe and application
A technology of cathepsin and agapanthus, applied in the fields of application, genetic engineering, plant gene improvement, etc., to achieve the effect of reducing cell proliferation rate and accelerating death
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1, the cloning of Agapanthus Abaciens ApCathB gene
[0053] 1. Acquisition of plant material
[0054] Take the leaf tissue of Agapanthus adult seedlings for RNA extraction;
[0055] 2. Extraction of RNA
[0056] Total RNA (Trizol: Invitrogen) was extracted with "RNA prep pure Plant Total RNA Extraction Kit", and the integrity of RNA was identified by formaldehyde denaturing gel electrophoresis, and then the purity and concentration;
[0057] 3. Full-length cloning of genes
[0058] According to the protein function annotation results of Agapanthus japonica transcriptome sequencing (RNA-seq), the core fragment of ApCathB gene of Agapanthus japonica was obtained. Using the RACE method (SMARTer TM RACE cDNA Amplification Kit: Clonetech) for full-length cDNA cloning in three stages:
[0059] (1) PCR to obtain the middle fragment of the gene
[0060] The extracted RNA was reverse-transcribed (Prime Script Ⅱ 1st Strand cDNA Synthesis Kit: Treasure Bioengi...
Embodiment 2
[0071] Example 2, Sequence Information and Homology Analysis of Agapanthus Abaciens ApCathB Gene
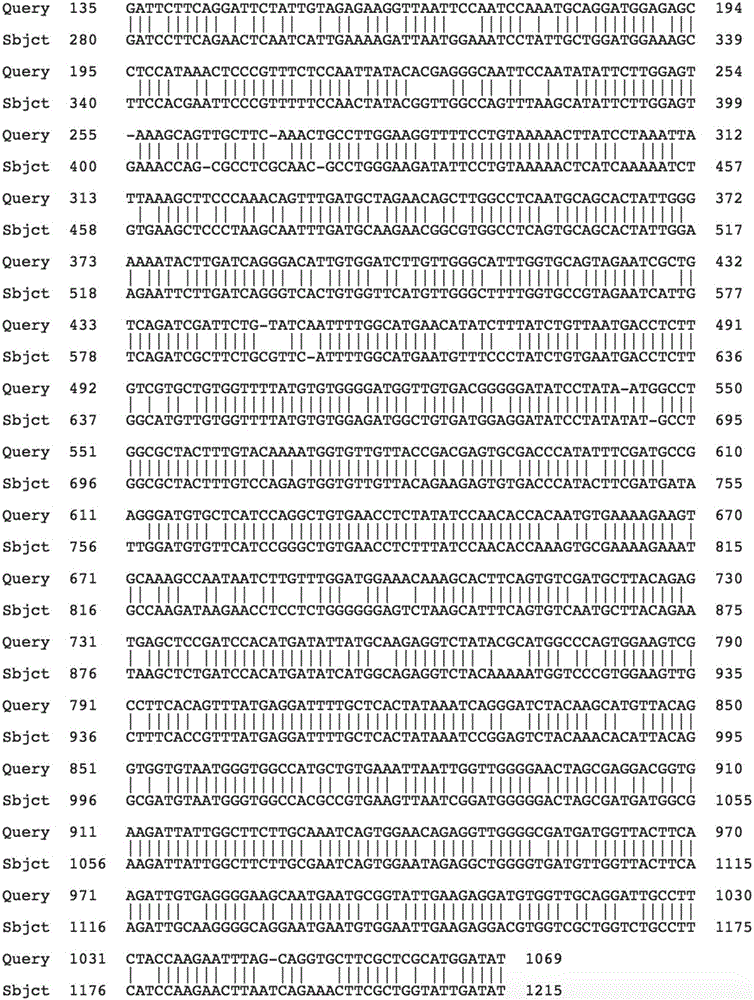
[0072] The full-length open reading frame sequence of the new Agapanthus agapanthus ApCathB gene of the present invention is 1071bp, and the detailed sequence is shown in the sequence shown in SEQID NO.3. According to the sequence of the open reading frame, the amino acid sequence of the Agapanthus Abaciens ApCathB protein was deduced, with a total of 356 amino acid residues, a molecular weight of 39.51kDa, and an isoelectric point (pI) of 5.64. For the detailed sequence, see the sequence shown in SEQ ID NO.4;
[0073] The open reading frame sequence of Agapanthus ApCathB and the amino acid sequence of its encoded protein were nucleated in Non-redundant GenBank+EMBL+DDBJ+PDB and Non-redundant GenBank CDS translations+PDB+SwissProt+Superdate+PIR databases using BLAST program Nucleotide and protein homology search, it was found that it has 81% identity with the oil palm Cathepsin...
Embodiment 3
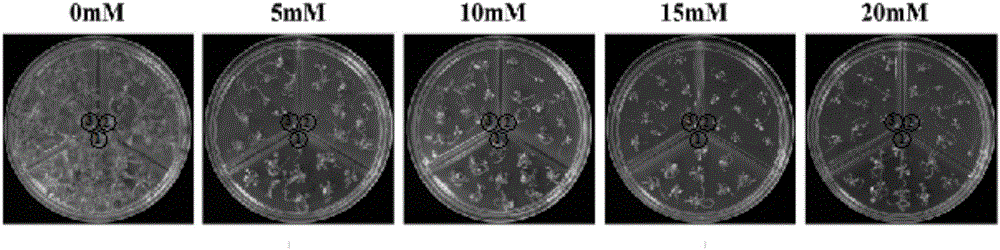
[0074] Embodiment 3, Agapanthus ApCathB gene transformation model plant Arabidopsis thaliana
[0075] 1. Construction of the expression vector containing the gene of interest (ApCathB)
[0076] According to the full-length coding sequence of Agapanthus ApCathB gene (SEQ ID NO.3), design primers to amplify the complete coding reading frame, and introduce restriction endonuclease sites on the upstream and downstream primers respectively (this can be determined by the selected vector Depends), in order to construct the expression vector. Using the amplified product obtained in Example 1 as a template, after PCR amplification, the coding region sequence of the ApCathB gene was connected to an intermediate vector (such as pMD19-T) for sequencing, and then the coding region of the sequenced correct ApCathB gene The sequence is further cloned into an expression vector (such as pHB), and transformed into Agrobacterium tumefaciens (such as GV3101) under the premise that the reading ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com