Detection primer set of Streptococcus agalactiae, detection kit and multiplex PCR detection method
A detection kit and technology for Streptococcus lactis, applied in biochemical equipment and methods, methods based on microorganisms, measurement/testing of microorganisms, etc., can solve the problem of loss of target fragments, missed or false detections, primer dimers, etc. problems, to reduce labor costs, improve quality and safety, and quickly detect pathogens
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Extraction of sample genomic DNA
[0041] 1. Extraction of genomic DNA from fish (including bacteria in fish):
[0042] 1) Take 50~100 mg of fish tissue to be tested and add 1000 μL of TE buffer solution, fully homogenize with a homogenizer, take 180 μL of tissue homogenate, transfer it to a 1.5ml centrifuge tube, add 20 μL of concentration Incubate at 30°C for 10 min with 50 mg / mL lysozyme.
[0043] 2) Add 10 μL of 10% SDS and 5 μL of 20 mg / mL proteinase K to the solution in step 1), and incubate at 37°C for 1 h.
[0044] 3) Add 50 μL of 5 mol / L NaCl solution to the solution after incubation in step 2), mix well, then add 40 μL of CTAB NaCl solution, and incubate at 65°C for 20 min.
[0045] 4) Add a mixed solution of phenol-chloroform-isoamyl alcohol equal to the volume of the incubated solution to the incubated solution in step 3), mix well, and centrifuge at 12000 g / min for 4-5 min.
[0046] 5) Transfer the supernatant after centrifugation in step 4) to...
Embodiment 2
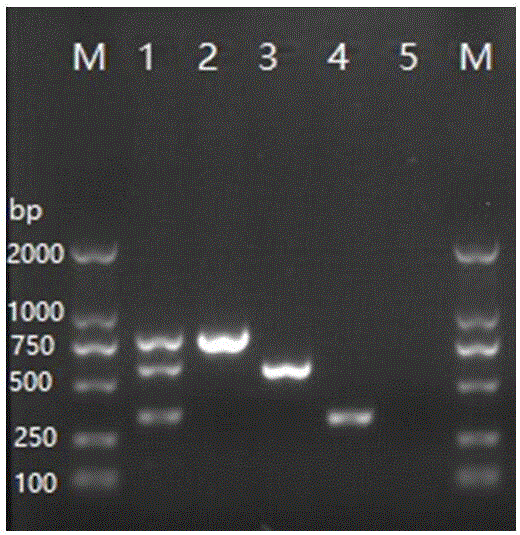
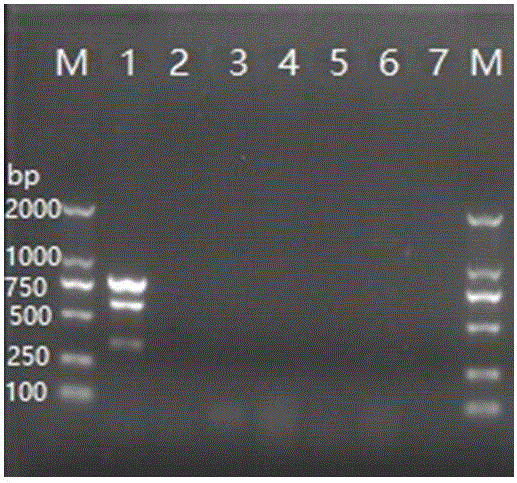
[0057] Example 2 Design and Effectiveness Detection of Streptococcus agalactiae Multiplex PCR Detection Primer Set
[0058] 1. Selection of three specific virulence genes of Streptococcus agalactiae hyB , pon A and cfb , using Primer Premier 6.0 software to analyze and design corresponding primer pairs, each primer pair can specifically identify the specificity of the above-mentioned bacterial virulence genes respectively, and the primer sequences are as follows:
[0059]
[0060] 2. Effectiveness testing:
[0061] (1) Synthesize the primer set designed above, and use the primers of the primer set to perform single PCR verification on the DNA of Streptococcus agalactiae extracted in Example 1, wherein the single PCR reaction system is as follows:
[0062]
[0063] (2) Simultaneously perform multiple PCR verification on the three virulence genes of Streptococcus agalactiae, using the following multiple PCR reaction system:
[0064]
[0065] (3) Use the following...
Embodiment 4
[0075] Example 4 Kit
[0076] The detection kit of this embodiment can be used for multiplex PCR to quickly detect whether the sample contains Streptococcus agalactiae, and the detection kit consists of Streptococcus agalactiae hyB , pon A and cfb Gene detection primer set, TE buffer, lysozyme, proteinase K, SDS solution, phenol-chloroform-isoamyl alcohol mixed solution, isopropanol, ethanol with a volume concentration of 70%, NaCl solution of CTAB, positive control substance and PCR DsMix consists of:
[0077] (1) Primer set for Streptococcus agalactiae detection: 1 tube containing Streptococcus agalactiae hyB Gene primers hylB-F and hylB-R, the nucleotide sequences of which are shown in SEQ ID NO.1 and SEQ ID NO.2 respectively; pon A Gene primers ponA-F and ponA-R, the nucleotide sequences of which are shown in SEQ ID NO.3 and SEQ ID NO.4 respectively; 3 tubes contain Streptococcus agalactiae cfb The nucleotide sequences of gene primers cfb-F and cfb-R are respectiv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com