A molecular marker related to milk production traits of dairy goats and its application
A technology of molecular markers and dairy goats, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., to achieve the effect of less equipment, speed up the breeding process, and shorten the generation interval
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1 Amplification of Partial Sequence of H-FABP Gene of Laoshan Dairy Goat
[0054] 1. Primer design
[0055] Using the online primer design software Primer-BLAST (https: / / www.ncbi.nlm.nih.gov / tools / primer-blast / ), the goat H-FABP gene sequence (GeneBank accession number: NC_030809.1, sequence interval 13664760- 13673332) was used as a template to design amplification primers containing partial sequences. The primers were synthesized by Qingdao Qingke Zixi Biotechnology Co., Ltd. The DNA sequences of the primer pairs are as follows:
[0056] H-FABPF5:TGTTCTGACCCCAAACTCGG
[0057] H-FABPR5:AGGGCTAGAGAACTGCTCCG
[0058] 2. Purification and sequencing of PCR products
[0059] PCR amplification reaction system 25 μL, including 1 μL of dairy goat peripheral blood genomic DNA (50ng / μL), 12.5 μL of 2×PCR Mix (Shanghai Sangon Bioengineering Co., Ltd.), 0.75 μL of forward primer H-FABPF4 (10mM ), 0.75 μL of reverse primer H-FABPR4 (10 mM), 10 μL of ddH 2 O. The react...
Embodiment 2
[0071] Example 2 Genotyping
[0072] The M of the 147th mutation site in the above sequence is A or C, and the mutation leads to SmaI-RFLP polymorphism. However, there is also a SmaI restriction site (CCCGGG, 165-170) near the mutation site at position 147 in this sequence, which makes it difficult to classify by restriction enzyme digestion, so another pair of primers (one of which is a mutation primer) was designed to solve the problem. For this difficulty, the sequence is as follows:
[0073] Sma1F:GGGACCAGGGGACGGATGTCG
[0074] Sma1R1:GCACCCAGTTTGCCTACGTCACCGCGGGTACCGGGCTCCGAGTCCCTGC
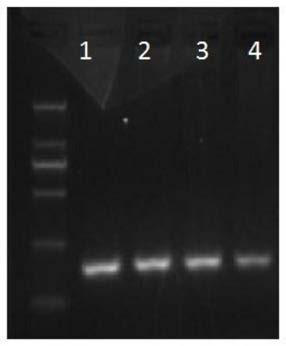
[0075] The sequence of the 176bp PCR product obtained by using the primer pair Sma1F and Sma1R1 amplified is as follows (the product was detected by electrophoresis on a 2% agarose gel, and the results are shown in figure 1 ):
[0076] GGGACCAGGGGACGGATGTCGGTGGCCTGCGCCCCAGCTCGCGGGCACCGTGCCCCCAAC
[0077] TGCTTCCTTTCCAGATTCCGAAGAGCGCCTTGAGGCCAGGGAAGGGAGCAGGGCTTACCA
[0078] GCCMGGGGCAGG...
Embodiment 3
[0080] Example 3 Association analysis of genotypes and traits of Laoshan dairy goat population
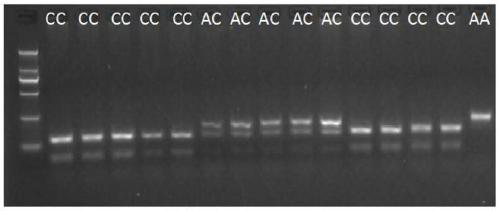
[0081] Select 200 Laoshan dairy goat ewes in lactation stage from Qingdao Aote sheep farm, collect whole blood from the jugular vein, extract genomic DNA, use primer pair Sma1F, Sma1R1, carry out PCR amplification on dairy goat population DNA, and use SmaⅠ enzyme digestion The genotype of each individual was detected, and it was found that there were 117 CC genotype individuals (group 1), and the average daily milk production measured was 2.32±0.68kg; the AC genotype individuals were 80 (group 2), and the daily milk production The average weight was 2.17±0.53kg; there were 3 individuals with AA genotype, because the number of individuals with AA genotype was too small to be statistically significant, so statistical analysis was not performed. Statistical analysis was performed on the data of group 1 and group 2 using one-way analysis of variance, and the obtained P value was 0.029 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com