Cascade enzyme reactor for enzymatically digesting DNA into mononucleoside
An enzyme reactor and DNase technology, applied in the field of DNase digestion, can solve the problems of complex enzymatic hydrolysis system, and achieve the effect of good stability and low back pressure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
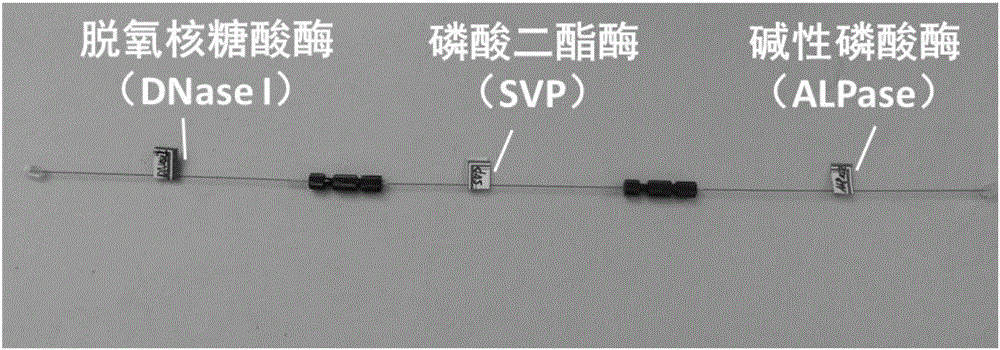
[0026] Prepare deoxyribonuclease, phosphodiesterase and alkaline phosphatase immobilized DNA single enzyme reactor respectively, and then these three enzyme reactors are in the order of deoxyribonuclease-phosphodiesterase-alkaline phosphatase Cascaded DNase reactors are obtained in series, such as figure 1 . Among them, deoxyribonuclease in the presence of Mg 2+ When present, it can digest single-stranded or double-stranded DNA to produce single-deoxynucleotides or oligodeoxynucleotides; phosphodiesterase can release 5'-mononucleotides from the 3'-terminus Nucleotides; and alkaline phosphatase can dephosphorylate single nucleotides to obtain single nucleoside products. The preparation method of single-enzyme enzyme reactor is as figure 2 ,Specific steps are as follows:
[0027] 1. Preparation of capillary silica gel monolithic column: several fused silica capillary tubes were cut, washed with water and methanol for 30 minutes each, then rinsed and activated with 1M NaOH s...
Embodiment example 2
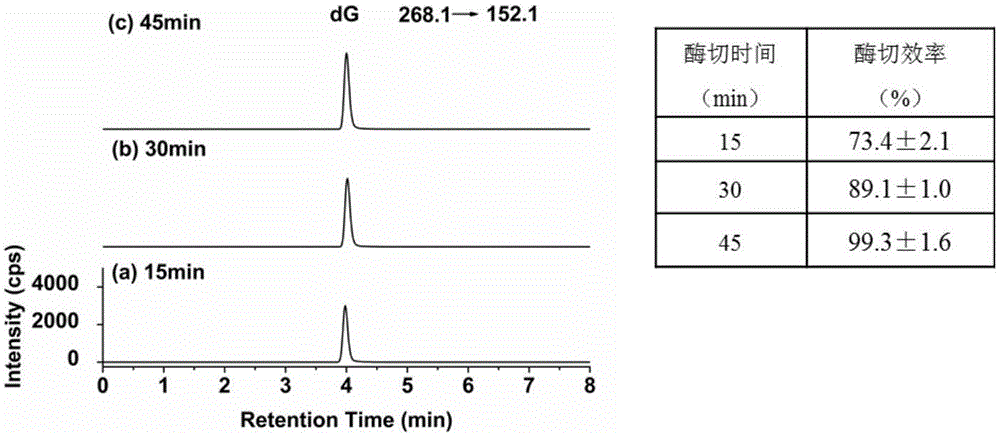
[0033] Cascade DNase Reactor Enzymatically digests calf thymus DNA for detection of deoxyguanosine (dG): inject 12 μL of 200 ng / μL calf thymus DNA into the cascade DNase reactor through a low-pressure micro-injection pump, and then use 10 mM Tris- HCl (pH=7.6, 2mM Mg 2+ , 2mM Ca 2+ ) to flush out the enzymatic hydrolysis solution, and digest at room temperature (25° C.) for 15, 30, and 45 minutes respectively until 60 μL of enzymatic hydrolysis samples are collected. LC-MS was used to quantitatively detect dG in the digestion product, and the chromatographic peaks under different digestion times were detected as image 3 shown.
[0034] Main parameter settings of the LC-MS used:
[0035] (1) Chromatographic column: Zorbax Eclipse Plus C18column (2.1×100mm, 1.8μm, Aglient)
[0036] (2) Mobile phase ratio: isocratic separation, 5.0% methanol and 95.0% water (0.1% formic acid)
[0037] (3) Flow rate: 0.3mL / min
[0038] (4) Column temperature: 30°C
[0039] (5) Injection vo...
Embodiment example 3
[0043] Cascade DNase reactor enzymatically digests calf thymus DNA for detection of dC: inject 12 μL of 200 ng / μL calf thymus DNA into the cascade DNase reactor through a low-pressure microsyringe pump, and then use 10 mM Tris-HCl (pH=7.6 , 2mM Mg 2+ , 2mM Ca 2+ ) to flush out the enzymatic hydrolysis solution, and digest at room temperature (25° C.) for 15, 30, and 45 minutes respectively until 60 μL of enzymatic hydrolysis samples are collected. LC-MS was used to quantitatively detect dC in the digestion product, and the chromatographic peaks under different digestion times were detected as Figure 4 shown.
[0044] The main parameter setting of the LC-MS used is the same as that of the implementation case 2, and the quantitative characteristic ion pair is set as: dC m / z 228.1→112.1 (5eV).
[0045] The experimental results show that with the increase of enzymatic hydrolysis time, from 15 minutes to 45 minutes, the content of dC obtained by enzymatic cascade DNase reaction...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com