HPV16E7 protein nano-antibody as well as preparation method and application thereof
A HPV16E7, 1.HPV16E7 technology, applied in the fields of botanical equipment and methods, biochemical equipment and methods, applications, etc., can solve the problems of poor antibody stability, high production cost, and limited papillomavirus detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1: Induced expression and purification of HPV16 E7 protein
[0034] (1) Induced expression of HPV16E7 protein
[0035] First, transform the successfully constructed pSUMO-HPV16E7 vector into BL21 (DE3) to obtain pSUMO-HPV16E7 recombinant bacteria, select a single clone in kanamycin liquid LB, and culture overnight; then inoculate at a ratio of 1:100 Incubate 1.5-2h in mycin liquid LB, when the bacteria liquid OD 600 When it is 0.6-0.8, induce expression with 23°C, 16h, 0.1mmol / L IPTG.
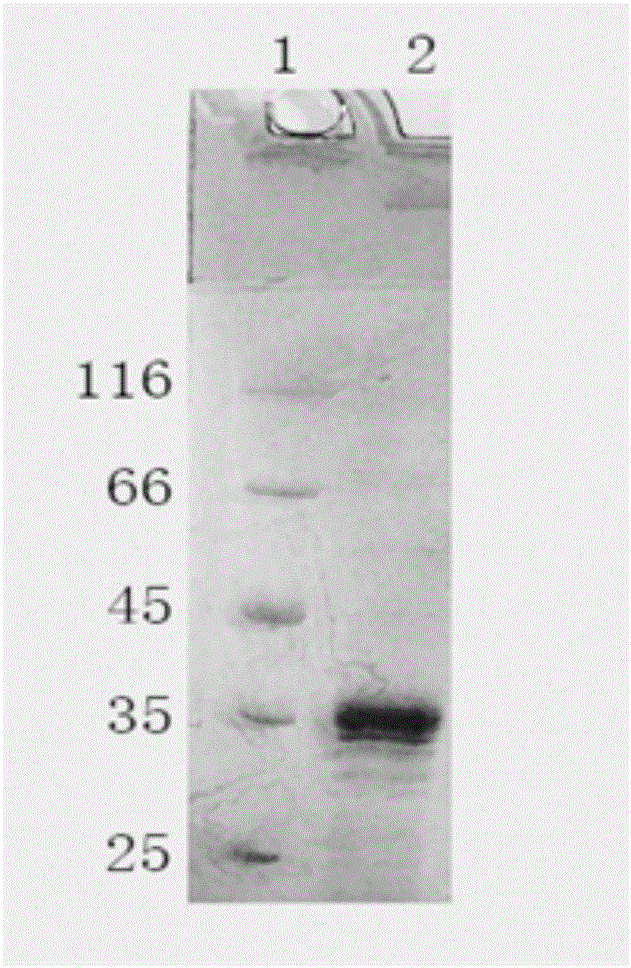
[0036] figure 1 SDS-PAGE electrophoresis detection results showed that the induced HPV16 E7 protein expression bacteria had a band at 35KD, which was in line with the expected molecular weight of the target protein.
[0037] (2) Purification of HPV16 E7 protein
[0038] Purified HPV16E7 protein was obtained by Ni-IDA affinity chromatography.
[0039] The chromatographic column was first washed with bacterial cell buffer, and Ni-IDA was added to fill the chromatographic c...
Embodiment 2
[0041] Embodiment 2: Construction of HPV16 E7 protein Nanobody library
[0042] (1) HPV16 E7 protein immunization of Bactrian camels in Xinjiang
[0043] Mix 1 mg of HPV16 E7 protein with an equal volume of Freund's adjuvant, immunize Xinjiang Bactrian camels (subcutaneous injection at 3-5 points), and immunize 7 times (complete Freund's adjuvant for the first time, and incomplete Freund's adjuvant for the rest). Adjuvant); After the immunization, 200ml of peripheral blood from the immunized camel was taken to separate and obtain peripheral blood lymphocytes.
[0044] (2) Separation of peripheral blood lymphocytes and extraction of total RNA
[0045]Firstly, Percoll density gradient centrifugation was used to separate and purify camel peripheral blood lymphocytes. After washing the lymphocytes with normal saline for several times, add Trizol and let it stand at room temperature for 10 minutes; then add chloroform to shake vigorously and let it stand at room temperature for 1...
Embodiment 3
[0079] Example 3: Panning HPV16E7 nanobody using nickel ion metal chelation affinity chromatography medium Ni-NTA.
[0080] (1) Cleaning of Ni-NTA medium:
[0081] Take 100ul of Ni-NTA medium in a 1.5ml EP tube, add 1ml of sterilized water, and mix with a vortex shaker; centrifuge at 3000rpm for 30s, discard the supernatant; wash 5 times in total, and replace the sterilized water with 0.05% TBST for the last time.
[0082] (2) closed:
[0083] Add 1ml of blocking solution (0.5% BSA) to the washed Ni-NTA medium, turn over and shake for 1 hour; after the blocking is completed, wash with 1ml TBST for 4 times.
[0084] (3) Negative screening to remove non-specifically bound phages:
[0085] The VHH-T7 phage library of HPV16E7 was diluted to 1ml with TBST, added to the blocked Ni-NTA medium, placed on an inversion shaker, and combined at room temperature for 30 minutes; centrifuged, and the supernatant was the T7 phage library after negative screening.
[0086] (4) Screening of ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com