Screening and application of a group of SNPs related to sheep wool traits
A sheep wool, sheep technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of high-throughput screening methods that are not widely developed, small in scope, and high in accuracy, and can reduce The cost of conventional breeding, the effect of speeding up the breeding process and reducing the degree of dependence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1 Skin tissue DNA extraction
[0018] (1) Fully shred 0.3 g sheep skin tissue sample with surgical scissors, place it in a 1.5 mL centrifuge tube, and add 500 μL tissue DNA extraction solution.
[0019] (2) Add RNase solution to the above centrifuge tube to a final concentration of 20 μg / mL, mix thoroughly, and digest in a 37°C water bath for 1 hour.
[0020] (3) Add proteinase K to the above centrifuge tube to a final concentration of 150 μg / mL, mix well, and digest in a water bath shaker at 55°C for 12-24 hours.
[0021] (4) Add an equal volume of Tris saturated phenol into the centrifuge tube, and slowly invert up and down until fully mixed. Centrifuge at 1200 rpm for 10 min at 4°C, then transfer the supernatant to a new centrifuge tube, and repeat step (4) once.
[0022] (5) Add an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1), mix well, and centrifuge at 1200 rpm for 10 min at 4°C.
[0023] (6) Transfer the supernatant to a new centrifuge...
Embodiment 2
[0028] Example 2 Genome PCR amplification
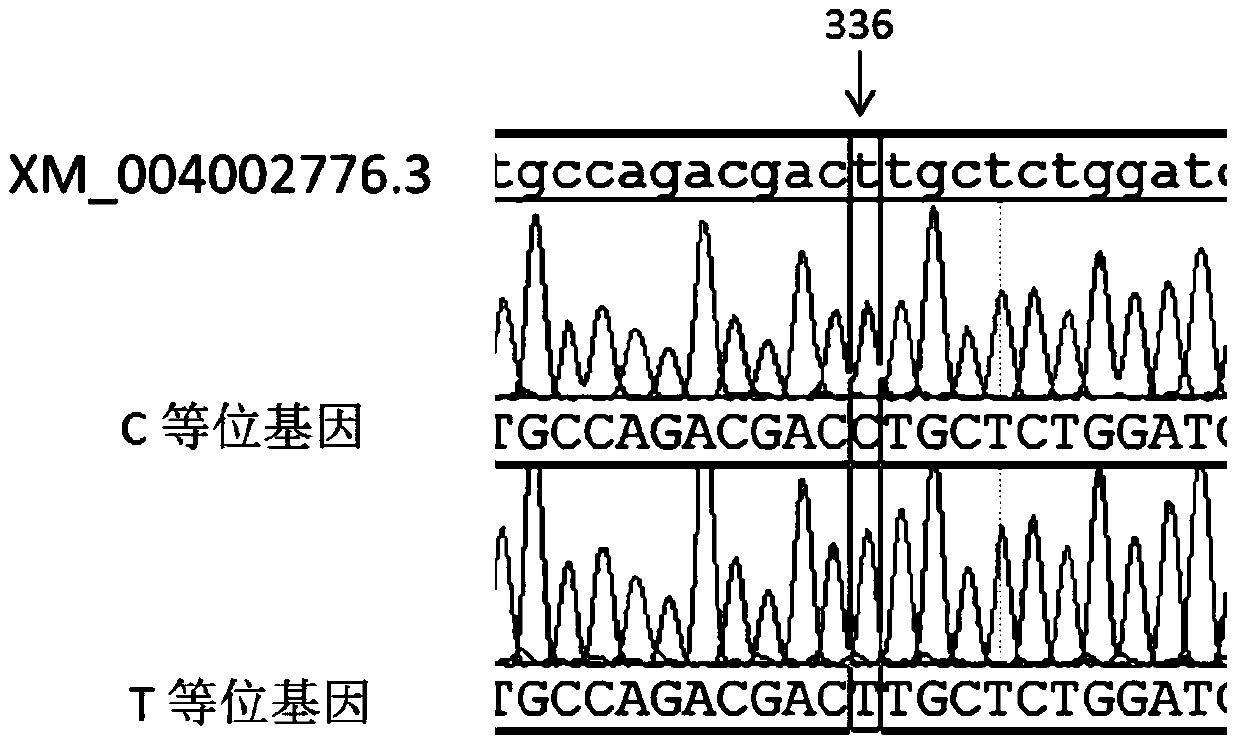
[0029] At present, there is no genome sequence information of sheep KAP13.1, and considering that the KAP family genes are single-exon genes, PCR-SSCP primers were designed using the predicted sequence of sheep KAP13-1-like mRNA (XM_004002776.3) as a template, primer information See the table below.
[0030] Table 1. 3 pairs of KAP13.1 gene-specific primer sequences, annealing temperature and target fragment length
[0031]
[0032] Add to PCR tube:
[0033]
[0034]
[0035] Setting of PCR amplification temperature:
[0036] Pre-denaturation at 95°C for 3 minutes
[0037] 35 cycles: Denaturation at 94°C, 30sec
[0038] See Table 1 for annealing temperature, 30sec
[0039] Extend 72℃, 30sec
[0040] Extend 72℃, 10min
[0041] Cool down to 12°C for storage
[0042] The results of PCR amplification were detected by 2% agarose gel electrophoresis. Electrophoresis detection showed that the KAP13.1 gene 2 primer PCR prod...
Embodiment 3
[0043] Embodiment 3 PCR product SSCP analysis
[0044] (1) Clean the glass plate with detergent, rinse with distilled water repeatedly and dry it for later use.
[0045] (2) Put the glass plate into the plastic frame, seal it with 2.5% agarose gel, prepare 12% polyacrylamide gel according to the standard formula, mix it well and pour it into the glass plate, insert the tooth comb and let it rest at room temperature for more than 30 minutes to make the gel The glue is fully solidified.
[0046] (3) After confirming that the gel is fully solidified, carefully pull out the tooth comb, wash the sampling hole with distilled water, dry it with absorbent paper, put the glass plate on the electrophoresis tank, add an appropriate amount of 1×TBE electrophoresis solution, and confirm that there is no leakage. Afterwards, 300V, 60mA pre-electrophoresis for about 10min.
[0047] (4) Take 2 μL of PCR product, add 8 μL of denaturing buffer, centrifuge briefly and then denature at 98°C for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com