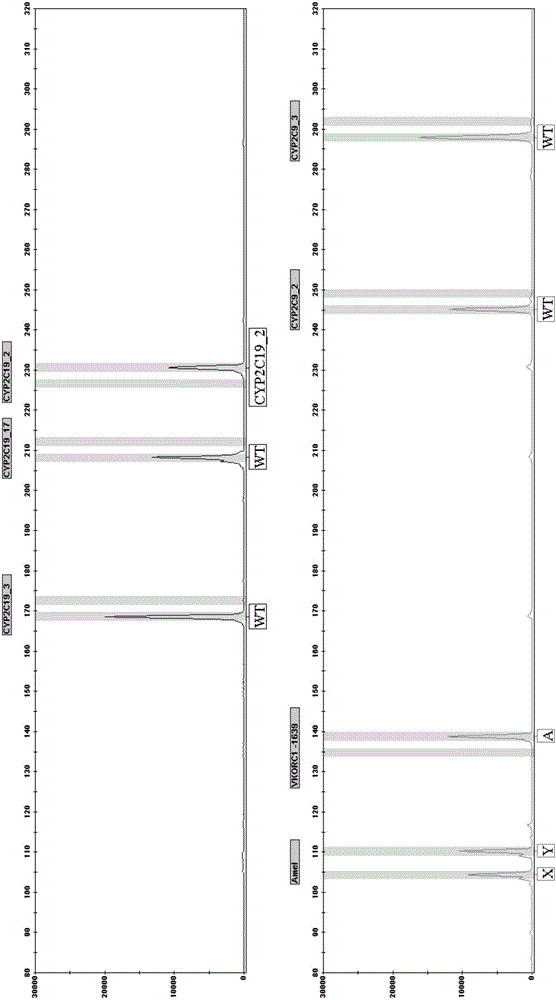
CYP2C19, CYP2C9 and VKORC1 genotyping multiplex amplification system and detection kit
A CYP2C19, 1.CYP2C19 technology, applied in the field of clinical molecular detection, can solve the problems of large template demand, high detection cost, and high demand for template quantity, and achieve the effect of high detection resolution and high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0098] Example 1: A kit for detecting CYP2C19, CYP2C9 and VKORC1 genotyping
[0099] This kit consists of PCR Master Mix, positive control DNA (normal person), negative control (sterilized water), and internal standard ROX500. The main components of PCR Master Mix include hot start DNA Taq enzyme (5U / μL, KAPA), UDG enzyme (5U / μL, NEB), 1.25×Buffer and primers for each site, etc. The dosage of each component is: when the reaction system is 20 μL, 1.25×Buffer is 16 μL, hot start DNA Taq enzyme is 0.4 μL, UDG enzyme is 0.04 μL, primer Mix is 0.58 μL (the final concentration of each primer
[0100] 100-200nM), template DNA or blood is 1-2μL, and sterilized water is added to 20μL.
[0101] The original primer sequences of the six SNP sites are as follows:
[0102] The base sequence of the primers at the 681G>A site of the CYP2C19*2 gene is as follows:
[0103] Forward wild-type primer: 5'-TTTCCCACTATCATTGATTATTTCCCG-3'
[0104] Forward mutant primer: 5'-TTTCCCACTATCATTGATTATT...
Embodiment 2
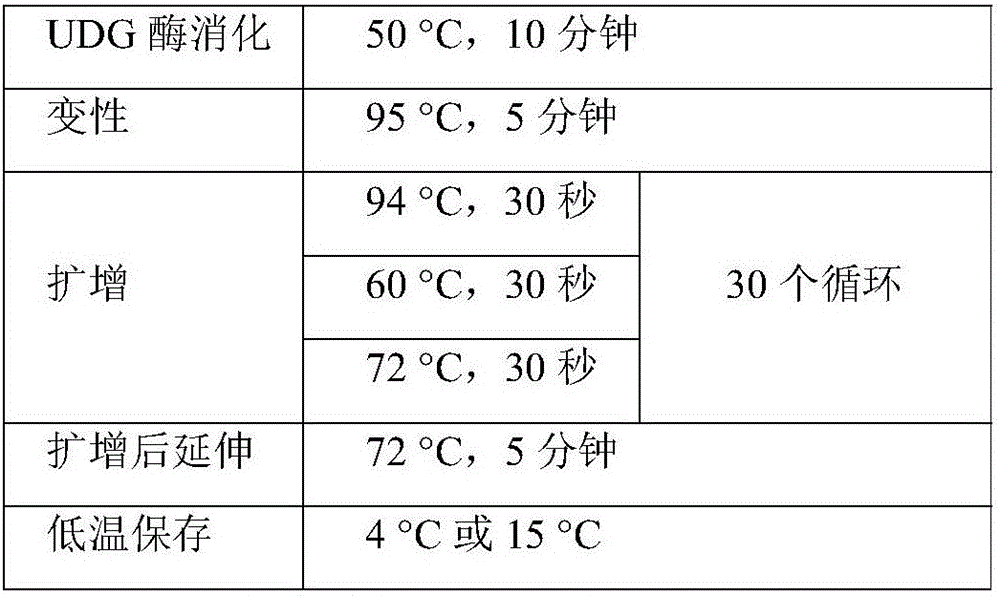
[0156] Example 2: Amplification method used in a kit for detecting CYP2C19, CYP2C9 and VKORC1 genotyping
[0157] Step 1: Extraction of Genomic DNA from Whole Blood
[0158] Take 200 μL of EDTA anticoagulated blood (sample A), and extract blood genomic DNA according to the instructions of the whole blood genomic DNA extraction kit. Measure the concentration of DNA with a spectrophotometer and dilute to 5-10ng / μL for use.
[0159] Step 2: PCR amplification reaction
[0160] 1. Aliquot PCR premix solution (completed in the reagent preparation area)
[0161] Shake and mix the PCR master mix solution (PCR Master Mix), and it is estimated that 4 detections will be performed, and each PCR reaction tube will be filled with 19 μL.
[0162] 2. Add template (completed in the specimen preparation area)
[0163] The detection templates are the blood and DNA of the above-mentioned patient A, and the templates, 1 μL of blood sample, 1 μL of DNA sample, 1 μL of positive control and 1 μL ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com