A microfluidic detection chip based on nucleic acid amplification technology and its preparation method
A detection chip and microfluidic technology, applied in biochemical equipment and methods, enzymology/microbiology devices, bioreactors/fermenters for specific purposes, etc. The method system is not perfect enough to achieve the effect of wide application, easy portability and small size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
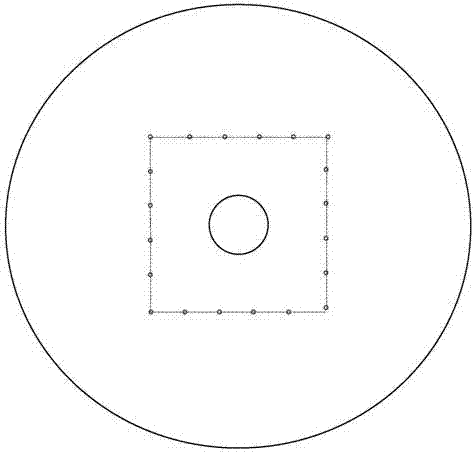
[0052] (1) Application: It is used for the detection of genetically modified ingredients in food. The target genes are endogenous gene ZSSIIb in corn, Lectin endogenous gene in soybean, SPS endogenous gene in rice, CAMV35S promoter, NOS terminator and BAR gene.
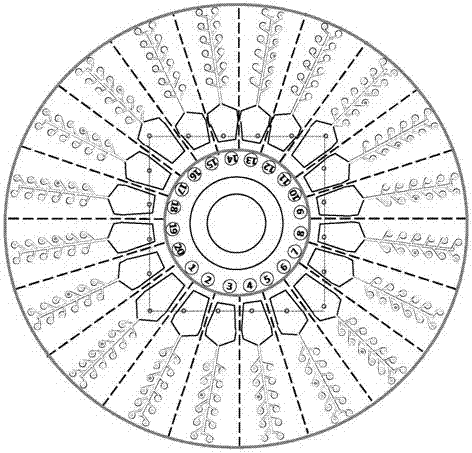
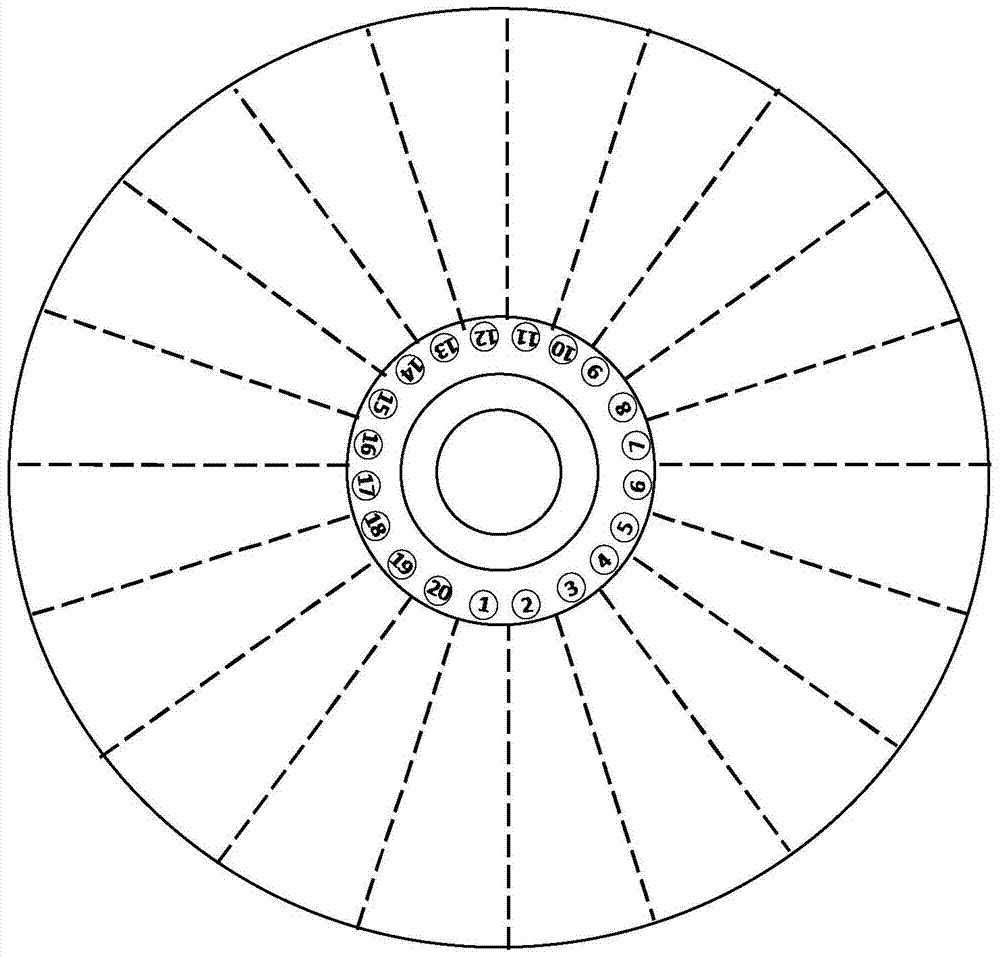
[0053] (2) Preparation process: Use computer software to draw the structure and channel graphics of each layer of the microfluidic detection chip for later inkjet printing, printing and etching; inkjet 20 serial number marking points and 20 on the bottom plate layer Reaction area: Apply printing technology on the primer layer to inkjet print oligonucleotide primer sequences in 20 reaction areas, 12 reaction chambers in each reaction area, a total of 240 areas, respectively endogenous gene ZSSIIb in corn, endogenous soybean Gene Lectin, rice endogenous gene SPS, CAMV35S promoter, NOS terminator and BAR gene, each primer was repeated twice, and stored in freeze-dry; 20 sample chambers, channels and 240 reaction chambers ...
Embodiment 2
[0058] (1) Application: It is used for the detection of animal-derived components in food. The targets are eukaryotic 18S, pigs, goats, sheep, cattle, buffaloes, chickens, ducks, geese, mice, foxes, and cats, a total of 12 targets.
[0059] (2) Preparation process: Use computer software to draw the structure and channel graphics of each layer of the microfluidic detection chip for later inkjet printing, printing and etching; inkjet 20 serial number marking points and 20 on the bottom plate layer Reaction area: Apply printing technology on the primer layer to inkjet print oligonucleotide primer sequences in 20 reaction areas, 12 reaction chambers in each reaction area, a total of 240 areas, respectively eukaryotic 18S, pig, goat, Sheep, cow, buffalo, chicken, duck, goose, mouse, fox, cat, 12 in total, freeze-dried and preserved; 20 sample chambers, channels and 240 reaction chambers were etched on the reaction layer, each reaction chamber volume was about 1 μL; print out the in...
Embodiment 3
[0064] (1) Application: used for the detection of foodborne pathogenic microorganisms in food, the targets are prokaryotic 16S, Escherichia coli, Salmonella, Staphylococcus aureus, Shigella, Listeria monocytogenes, a total of 6 .
[0065] (2) Preparation process: Use computer software to draw the structure and channel graphics of each layer of the microfluidic detection chip for later inkjet printing, printing and etching; inkjet 20 serial number marking points and 20 on the bottom plate layer Reaction area: Apply printing technology on the primer layer to inkjet print oligonucleotide primer sequences in 20 reaction areas, 12 reaction chambers in each reaction area, a total of 240 areas, respectively prokaryotic 16S, Escherichia coli, Salmonella, 6 sets of Staphylococcus aureus, Shigella, and Listeria monocytogenes, 2 replicates of each primer, freeze-dried and stored; 20 sample chambers, channels, and 240 reactions were etched on the reaction layer The volume of each reactio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com