Evolved immunoglobulin binding molecule D-C-G3 and preparation method and application thereof
A technology of D-C-G3 and immunoglobulin, applied in the field of protein, can solve the problems of high production cost, broad spectrum of IBPs and low binding force, and achieve the effect of improving sensitivity and accuracy, low cost and good effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Obtaining D-C-G3 molecules using molecular evolution methods
[0033] That is, a random combinatorial library of 7 single-binding domains including A, B, C, D, and E of SpA and B2 and B3 of SpG was constructed by phage display. Mouse IgG1, 2a, 2b, 3 and 8 kinds of IgG including human IgG, rabbit IgG, bovine IgG, and goat IgG were used as inducing molecules. Through the repeated process of "adsorption"-"elution"-"amplification", the The library undergoes 3-4 rounds of screening, and the positive clones that appear in each round of screening are sequenced to obtain novel combined sequences with advantageous binding properties.
[0034] 1. Construction of phage display library with seven single-domain random combinations of SpA and SpG
[0035]Using pCANTAB5S-SpA phagemid as a template, using uA and dAD, uB and dB, uC and dC, uD and dAD, uE and dE as upstream and downstream primers, PCR amplified A, B, C, D, E of SpA Five-domain fragments; using uG2 and dG2 as ...
Embodiment 2
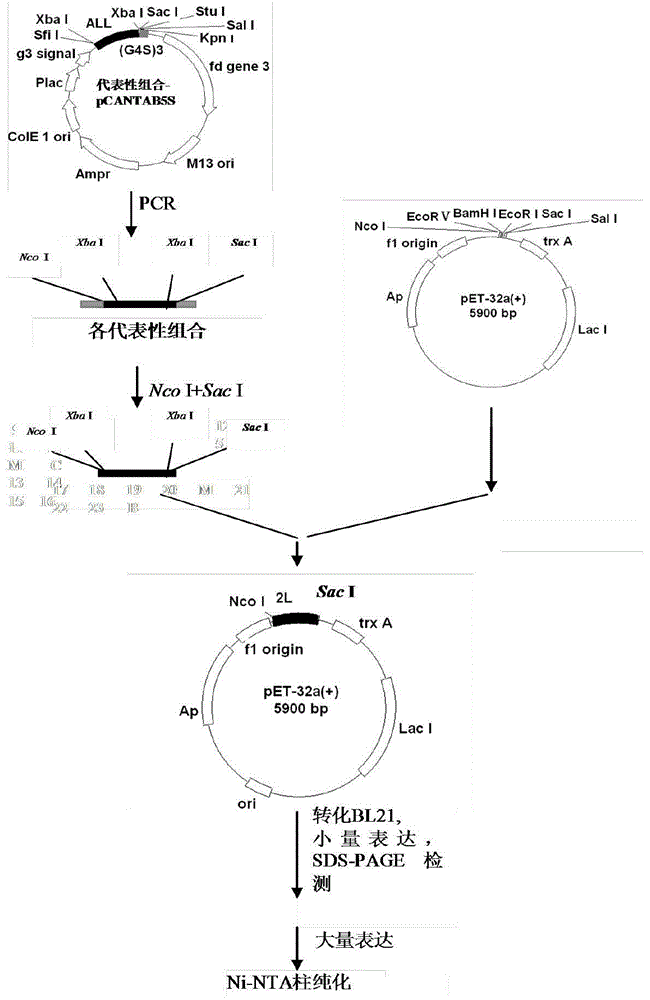
[0045] For the construction procedure of the prokaryotic expression vector pET32a(+)-D-C-G3, see image 3 .
[0046] 2.1 Primer synthesis
[0047] The primers used for PCR amplification of the cDNA sequence of the evolved immunoglobulin binding molecule D-C-G3 were: upstream 5SNco-u: 5’-GCGCCCATGGGGTCTAGAGCTGATGCGCAAC-3’, downstream 5SSac-d: 5’-GCGCCTCGAGTTATTCTAGACTGCTGGTGTTCGG-3’. The primer DNA sequence was synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0048] 2.2 PCR amplification of the expression sequence of D-C-G3
[0049] Using pCANTAB5S-D-C-G3 as a template, use the synthetic upstream and downstream primers to amplify, the reaction volume of PCR is 50 μl, add 5 ng of plasmid template, 1 μmol of upstream and downstream primers, 100 μmol of dNTP, Mg 2+ 3mmol, Taq enzyme 1U, add ddH 2 0 to 50 μl. Reaction conditions: 94°C for 30s-60°C for 30s-72°C for 45s; 35 cycles, 72°C extension for 5 minutes before the end of the reaction, the produ...
Embodiment 3
[0065] Example 3 Analysis of the binding characteristics of the novel immunoglobulin evolution molecule D-C-G3 to the four subclasses of mouse IgG, human IgG, rabbit IgG, bovine IgG and goat IgG
[0066] 1. Binding to the four subclasses of mouse IgG:
[0067] 1.1 Four subclasses of mouse IgG were purchased from Sigma, and SpA and SpG were purchased from Sigma.
[0068] 1.21 mg / ml human polyclonal IgG was dialyzed against PBS (pH 7.2). Take 50 μL of 3 mg / ml long-arm activated biotin (purchased from PIERCE company) and add 1 mL IgG (1 mg / mL), shake slightly at room temperature for 4 hours, then add a dialysis bag and dialyze overnight in PBS at 4 ° C. After the dialysis, add an equal amount of glycerol to - Store at 20°C.
[0069] 1. Adjust the concentration of 3D-C-G3, SpA and SpG to 1 mg / ml, and then dilute it with carbonate buffer (pH9.6) 1:200 to coat a 96-well plate. Each protein is coated with 3 rows of duplicate wells, at 4°C After 24 hours, wash with PBST 4-5 times, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com