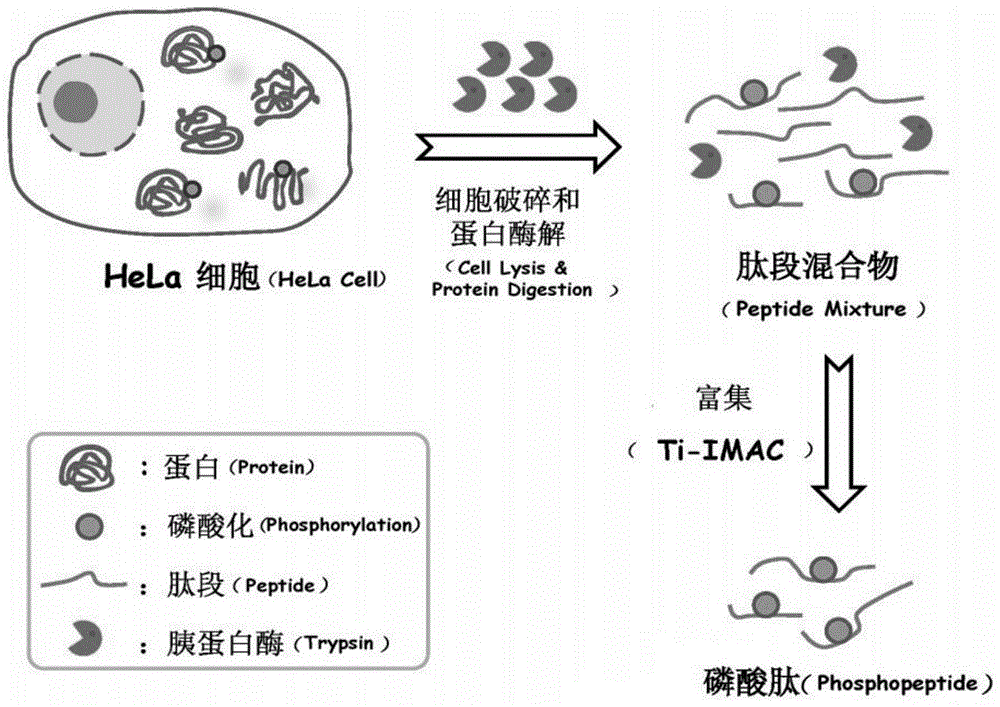
A rapid processing method for phosphorylated proteome samples
A technology for phosphorylating proteins and processing methods, applied in the preparation of test samples, etc., can solve the problems of high non-specific adsorption performance of immobilized enzymes, inconvenient operation, and no generalization, and achieve high-quality spectral identification coverage. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
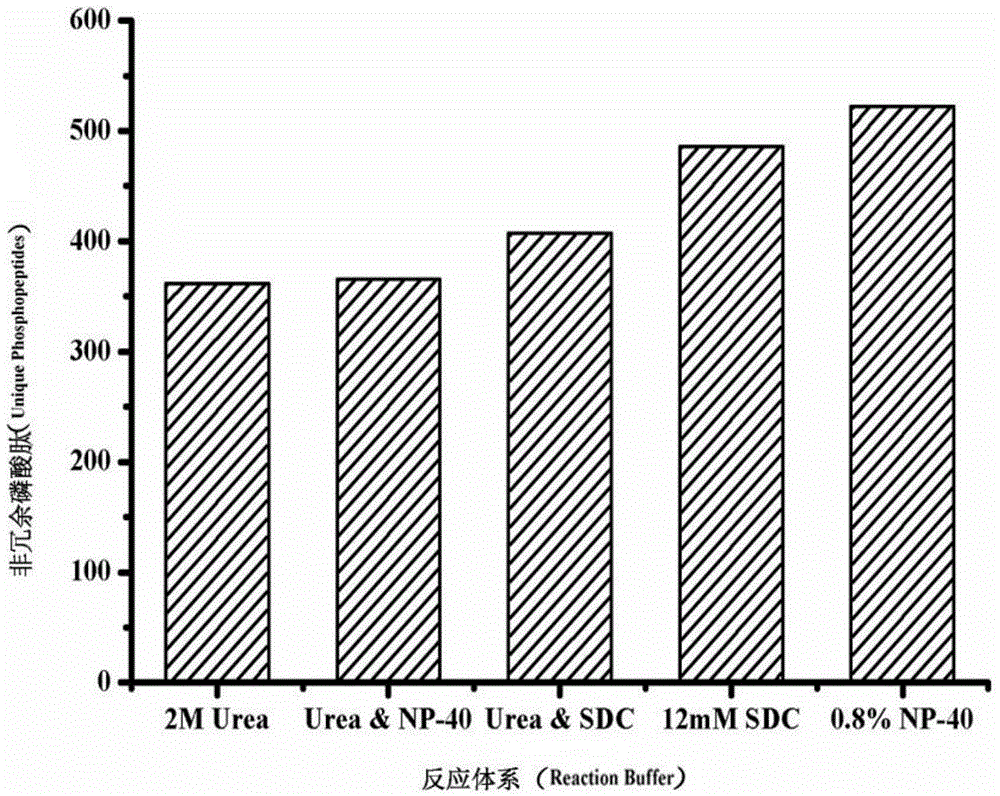
[0030] Investigation of the reaction system of cell lysis-proteolysis: 100,000 HeLa cell samples were placed in the following five reaction systems, respectively: 1) 2M urea buffer solution; 2) 0.8% NP-40 buffer solution; 3) 12mM sodiumdeoxycholate ( SDC) buffer solution; 4) 2M urea and 0.8% NP-40 buffer solution; 5) 2M urea and 12mM SDC buffer solution, these five buffer solutions contain 10mM DTT and phosphatase inhibitor respectively (ie 1mM NaF and 1mM Na3VO4) , then add 300ng / μL trypsin to the solution, vortex and shake for tens of seconds on the shaker until all the cell membranes are broken, then place the mixture in a 37°C water bath ultrasonicator for 1h, and finally, through a controllable centrifugal force And the GELoader Tip centrifuge filled with 1 mg Ti-IMAC material was used for enrichment of phosphopeptides, and the obtained phosphopeptides were lyophilized at room temperature, and redissolved in 100 μL 0.1% (v / v) formic acid for RP LC-MS / MS analysis , qualita...
Embodiment 2
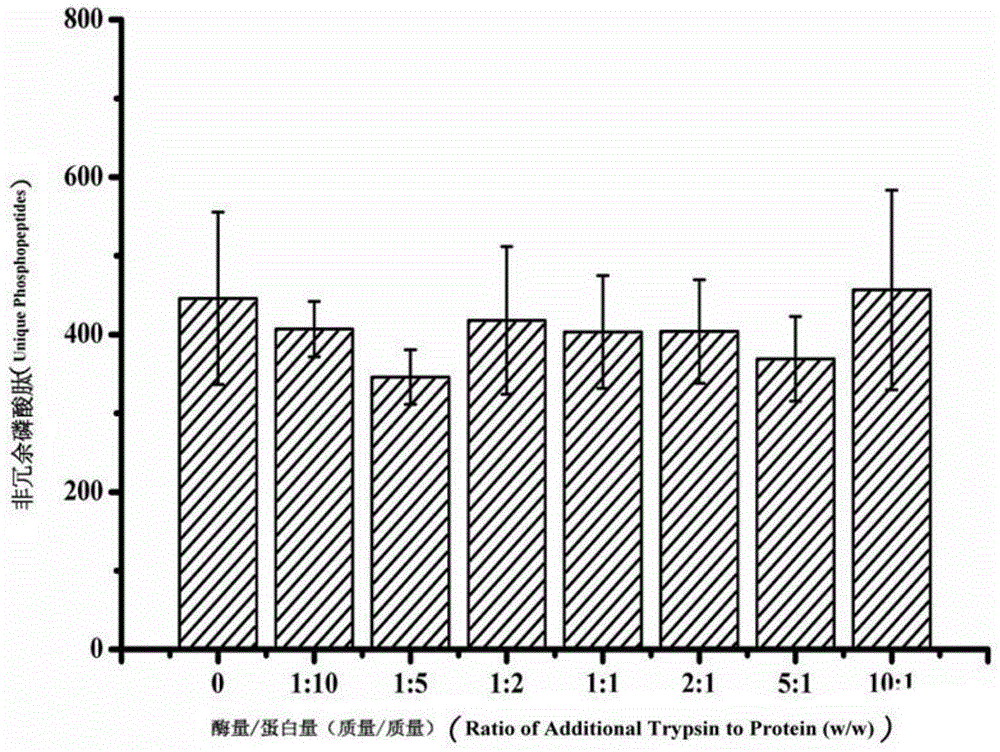
[0033] The effect of high-concentration trypsin on the enrichment of phosphopeptides and identification by mass spectrometry: Add a series of trypsin with different enzyme / protein ratios to 200 μg of HeLa hydrolyzate, mix well, and enrich with 2 mg of Ti-IMAC material. The enrichment steps of phosphopeptides are as follows: After resuspending the material with 80% ACN6%TFA, mix it with the enzymatic hydrolysis solution at a volume ratio of 1:1, shake at room temperature for 30 minutes, and after fully enriched, at 13500rpm Centrifuge for 3 minutes, discard the supernatant, then wash with 50% ACN6%TFA, 200mM NaCl and 30% ACN6%TFA in sequence, shake for 30 minutes, remove the supernatant by high-speed centrifugation as above, and finally wash with 10% NH 3 ·H 2 The phosphopeptide was eluted twice with O solution and subsequently identified by mass spectrometry as described in Example 1.
[0034] Such as image 3 As shown, the numbers on the axis of abscissa represent the exces...
Embodiment 3
[0036] Optimization of the reaction conditions for the rapid processing method of phosphorylated proteome samples: the optimal trypsin concentration and reaction time required for the reaction were investigated respectively. Add four groups of 100000 HeLa cell samples into 50 μL 0.8% NP-40 reaction system, then add 20ng / μL, 100ng / μL, 300ng / μL, 1.0μg / μL trypsin respectively, and vortex the solution on the shaker Rotate and oscillate for tens of seconds until all the cell membranes are broken, then place the mixture solution in a 37°C water bath ultrasonicator and incubate for 1 hour, and finally, use a GELoader Tip centrifuge with controllable centrifugal force and filled with 1mg Ti-IMAC material for phosphoric acid Peptide enrichment, as described in Example 1, was followed by mass spectrometric identification.
[0037] As mentioned above, after the optimal concentration of trypsin was investigated, four groups of 100,000 HeLa cell samples were added to the reaction system of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com