Neutral phytase mutant and application thereof
A technology of phytase and mutants, applied in the field of genetic engineering, can solve problems such as unfavorable acid phytase action, achieve the effects of reducing breeding costs, reducing pollution, and improving enzyme activity levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment 1 The acquisition of phytase gene
[0019] According to the gene sequence in the public gene database, the codon of the synthetic gene was optimized and the phytase gene PHYA (sequence is SEQ ID NO: 2) derived from Aspergillus niger was artificially synthesized, and the amino acid sequence encoded by it was SEQ ID NO: 2. ID NO: 1.
[0020] PCR primers and reaction conditions are as follows:
[0021] Primer 1 (F): GCGCGAATTCATGAGCTTCCGGTCCCTG
[0022] Primer 2 (R): TAAAGCGGCCGCTTAGGCGAAGCACTCGGC
[0023] The reaction conditions were: denaturation at 94°C for 5 minutes; then denaturation at 94°C for 30 seconds, renaturation at 56°C for 30 seconds, extension at 72°C for 1.5 minutes, and after 30 cycles, incubation at 72°C for 10 minutes. The results of agarose electrophoresis showed that the size of the PHYA gene was 1395bp.
Embodiment 2
[0024] Example 2 Construction and Screening of Phytase Mutants
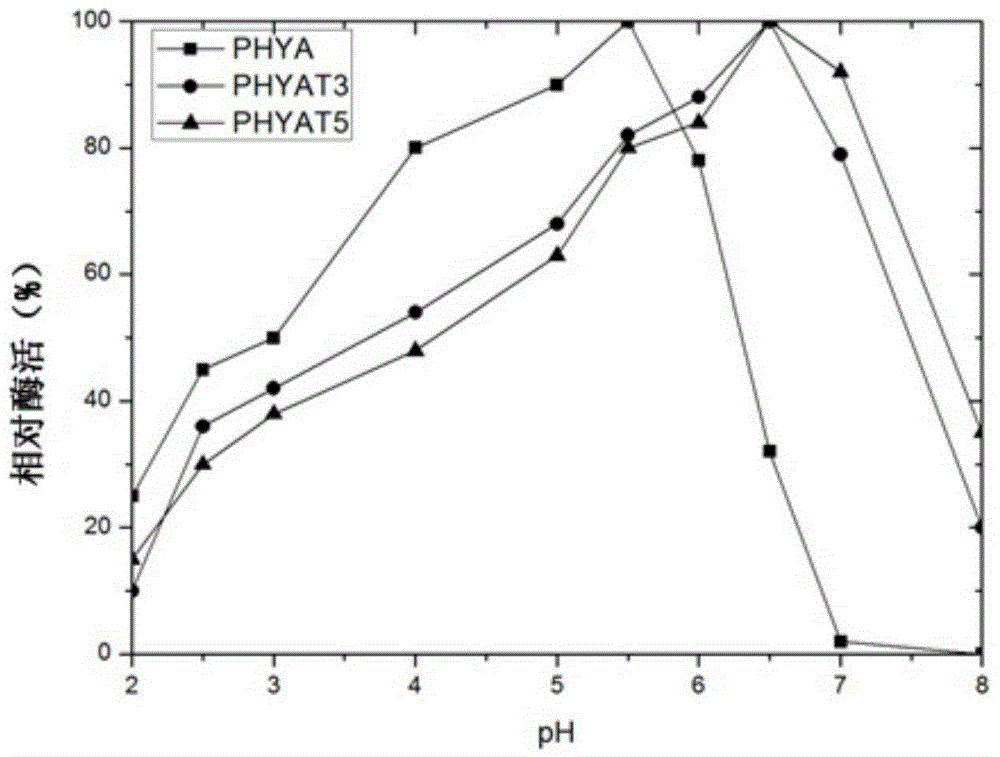
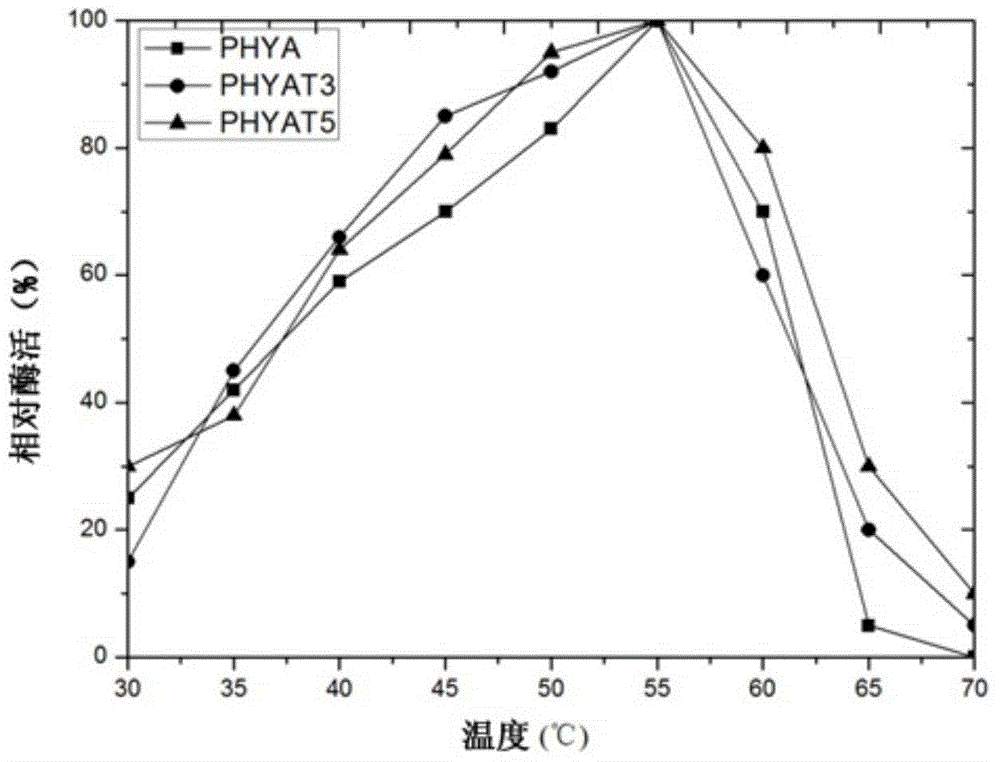
[0025] In order to improve the enzyme activity level of phytase PHYA under neutral conditions, the applicant screened a large number of mutations of the enzyme by directed evolution technology.
[0026] With the PHYA gene as a template, the primers 1 and 2 described in Example 1 were used for PCR amplification with the GeneMorph II random mutation PCR kit (Stratagene), and the PCR product was recovered by gel, and EcoRI and NotI were digested with the same enzyme. The final pET21a vector was ligated, transformed into Escherichia coli BL21(DE3), spread on LB+Amp plate, and cultured upside down at 37°C. After the transformants appeared, pick them one by one to a 96-well plate with a toothpick, and add 150ul to each well LB+Amp medium containing 0.1mM IPTG, cultivated at 37°C and 220rpm for about 6 hours, centrifuged to discard the supernatant, resuspended the bacteria with pH 5.0 buffer, and repeatedly freeze-thawe...
Embodiment 3
[0030] Example 3 Construction of recombinant expression vector
[0031] The gene fragments of phytase mutants PHYAT3 and PHYAT5 were amplified by PCR, and XbaI sites were introduced at both ends of the primers. The primer sequences are as follows:
[0032] Primer 3 (F): GC TCTAGA ATGAGCTTCCGGTCCCTG
[0033] Primer 4 (R): GC TCTAGA TTAGGCGAAGCACTCGGC
[0034] The PCR reaction conditions were as follows: denaturation at 94°C for 5 minutes; then denaturation at 94°C for 30 seconds, renaturation at 56°C for 30 seconds, extension at 72°C for 1.5 minutes, and after 30 cycles, incubation at 72°C for 10 minutes. The results of agarose gel electrophoresis showed that the PHYAT3 and PHYAT5 genes were fragments with a size of 1395 bp.
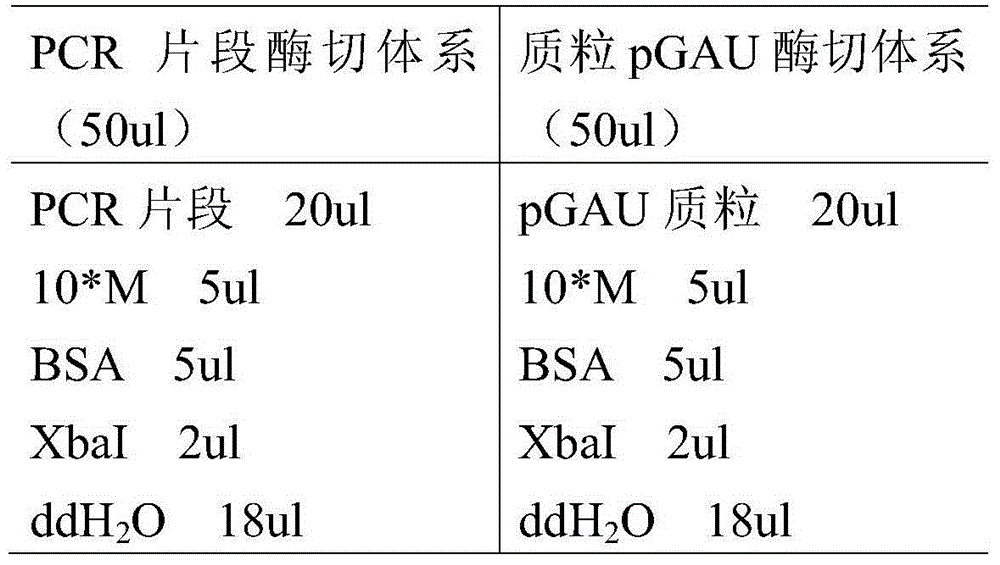
[0035] The phytase mutant PHYAT3 and PHYAT5 gene fragments obtained above and the expression vector pGAU were subjected to single-digestion with restriction endonuclease XbaI respectively, and the digestion conditions were as follows:
[0036] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com