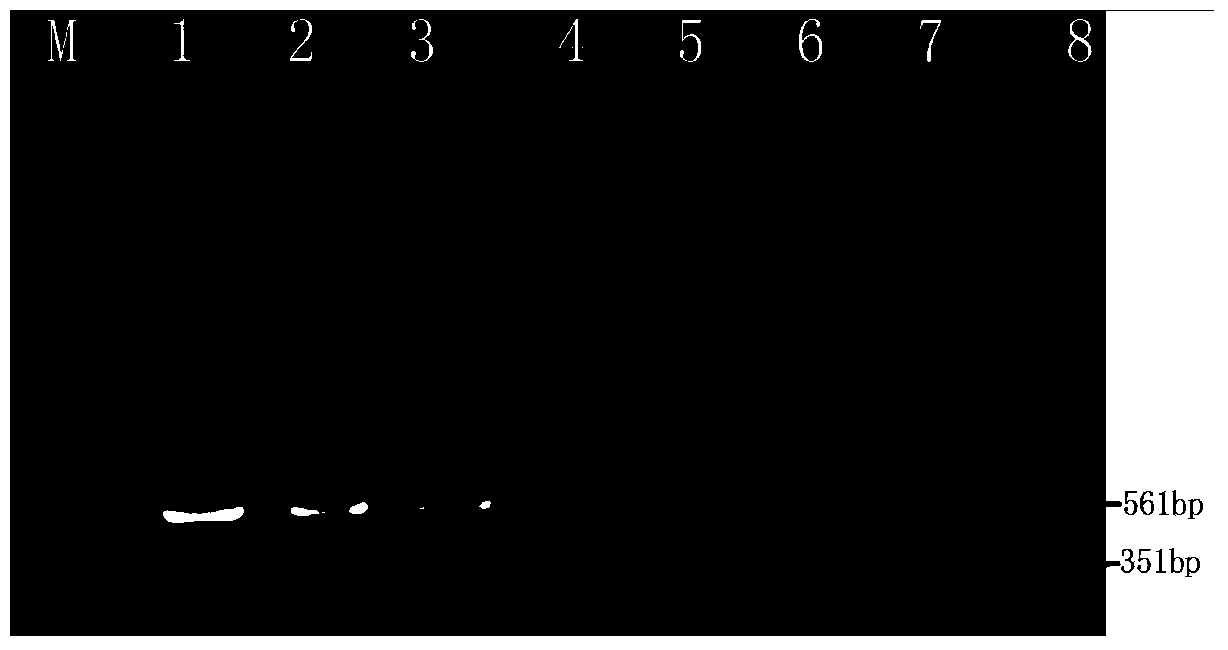Dual-PCR detecting method for pathogenic enterocolitis yersinia
A technology for enterocolitis and Yersinia, which is applied in the field of food safety detection, can solve the problems of instability, poor specificity, cumbersome operation, etc., and achieve the effect of strong specificity, high sensitivity, simple and fast operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1 Establishment of a dual PCR detection system for pathogenic Yersinia enterocolitica
[0020] 1. Extraction of genome and plasmid DNA: Inoculate Yersinia enterocolitica isolated by GB / T4789.8-2008 method into sterilized fresh soybean tryptone broth, cultivate overnight at 25-28°C, Take 1ml of bacterial solution or different dilutions of bacterial solution (6.5×10 9 -6.5×10 0 cfu / mL) Use the Genomic DNA Extraction Kit and Plasmid Extraction Kit of Guangzhou Meiji Biological Co., Ltd. to extract the genomic DNA and plasmid DNA of the pathogenic bacteria respectively, and store them at -30°C for later use;
[0021] 2. Double PCR amplification procedure:
[0022] PCR system (25 μL): 0.5-1.0 μL primers (the original primer concentration is 10 μM), including ail gene (its nucleotide sequence is as SEQ ID NO.1) primer and virF gene (its nucleotide sequence is as SEQ ID NO. 2) primers, wherein, ail gene primer: F: 5'-taatgtgtacgctgcgag-3' (its nucleotide sequence is...
Embodiment 2
[0028] Example 2 Application test of double PCR sensitivity detection of pathogenic Yersinia enterocolitica
[0029] Add 1 gram of yogurt (negative for pathogenic Yersinia enterocolitica) into 8ml sterile water, and then dilute with different dilutions (6.5×10 6 cfu / mL, 6.5×10 5 cfu / mL, 6.5×10 4 cfu / mL, 6.5×10 3 cfu / mL, 6.5×10 2 cfu / mL, 6.5×10 1 cfu / mL, 6.5×10 0 cfu / mL) of pathogenic Yersinia enterocolitica. Aseptically pipette 1.0ml of the bacterial solution to extract the genome and plasmid DNA of the pathogenic bacteria using a genome extraction kit and a plasmid extraction kit, respectively.
[0030] PCR system (25 μL): 1.0 μL primers (original primer concentration 10 μM) (ail gene: F: 5'-taatgtgtacgctgcgag-3', R: '-gacgtcttacttgcactg-3'; virF gene: F: 5'-ggcagaaca gcagtcagacata-3 ', R: 5'-ggtgagcatagagaatacgtcg-3'), 200-250μM dNTP, Ta q DNA polymerase 4U, 10× buffer 2.5μL, 5ng DNA template, add double distilled water to make up to 25μL.
[0031] Double PCR amplifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com