DNA (Deoxyribose Nucleic Acid) molecule, pichia pastoris recombinant plasmid and pichia pastoris recombinant bacterium for efficiently expressing PprI protein of deinococcus radiodurans
A DNA molecule, the technology of Pichia pastoris, applied in the field of Pichia pastoris recombinant bacteria, can solve the problems of reduced activity, unfavorable high expression of radioduran PprI protein, and different amino acid codon preferences of composition and function proteins.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Embodiment 1: Optimization and synthesis of DNA molecule of the present invention
[0050] On the premise of keeping the amino acid sequence of PprI protein unchanged, the present invention optimizes and transforms the sequence of the pprI gene (DR_0167, Gene ID: 1798483) of Deinococcus radiodurans R1 (Deinococcus radiodurans R1) open reading frame (Open Reading Frame, ORF), and encodes and synthesizes A new pprI gene—Pi-pprI gene, which is the nucleotide sequence shown in SEQ ID NO:1, was developed in order to efficiently express the target protein.
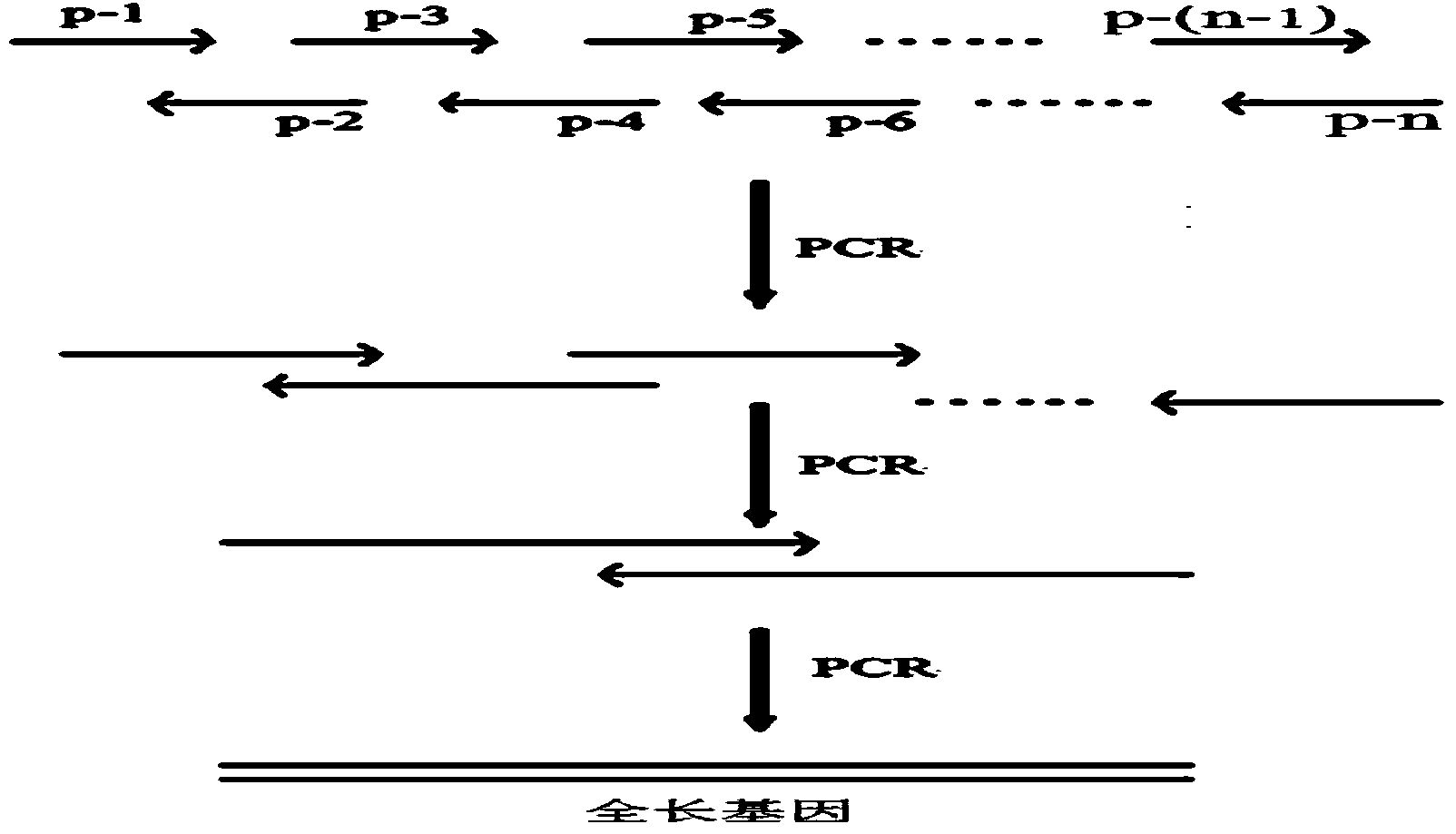
[0051] 1. Synthesis method
[0052] According to the artificially designed Pi-pprI gene, the present invention designs and synthesizes a series of overlapping (OVERLAP) primers of the nucleotide sequence shown in SEQ ID NO: 2-41, and synthesizes the primers shown in SEQ ID NO: 1 through Overlapping PCR. Nucleotide sequence and DNA molecules with CpoI restriction endonuclease sites and NotI restriction endonuclease sites ...
Embodiment 2
[0059] Example 2: Synthesis of DNA molecules (introduced with 6×His tag sequence) according to the present invention
[0060]Using the DNA molecule obtained in Example 1 as a template, use a primer with a nucleotide sequence shown in SEQ ID NO:42 and a primer with a nucleotide sequence shown in SEQ ID NO:41 with a 6×His tag sequence (CATCATCACCACCATCAT) Perform PCR amplification to obtain a DNA molecule (with a CpoI restriction endonuclease site and a NotI restriction endonuclease site) comprising the nucleotide sequence shown in SEQ ID NO: 1 and a 6×His tag sequence. Wherein, the primer SEQID NO:42 introduces a 6×His tag sequence based on the primer p-1.
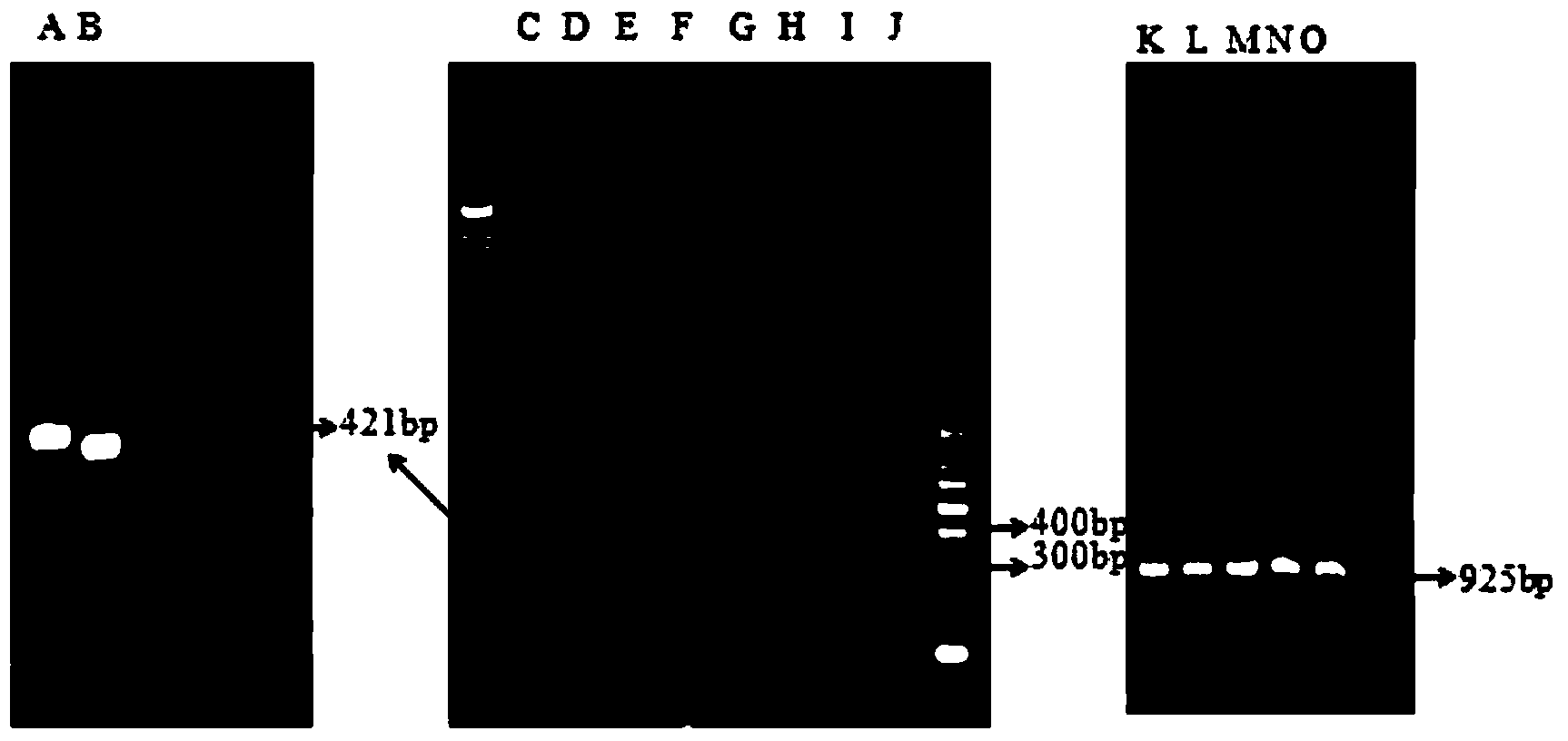
[0061] PCR amplification conditions were as follows: pre-denaturation at 95°C for 5 min, denaturation at 94°C for 30 s, annealing at 50°C for 30 s, extension at 72°C for 90 s, and a total of 30 cycles, and finally extension at 72°C for 10 min, and incubation at 4°C.
[0062] Get 3 μ l of PCR amplification product and detec...
Embodiment 3
[0063] Embodiment 3: Construction of Pichia pastoris recombinant plasmid
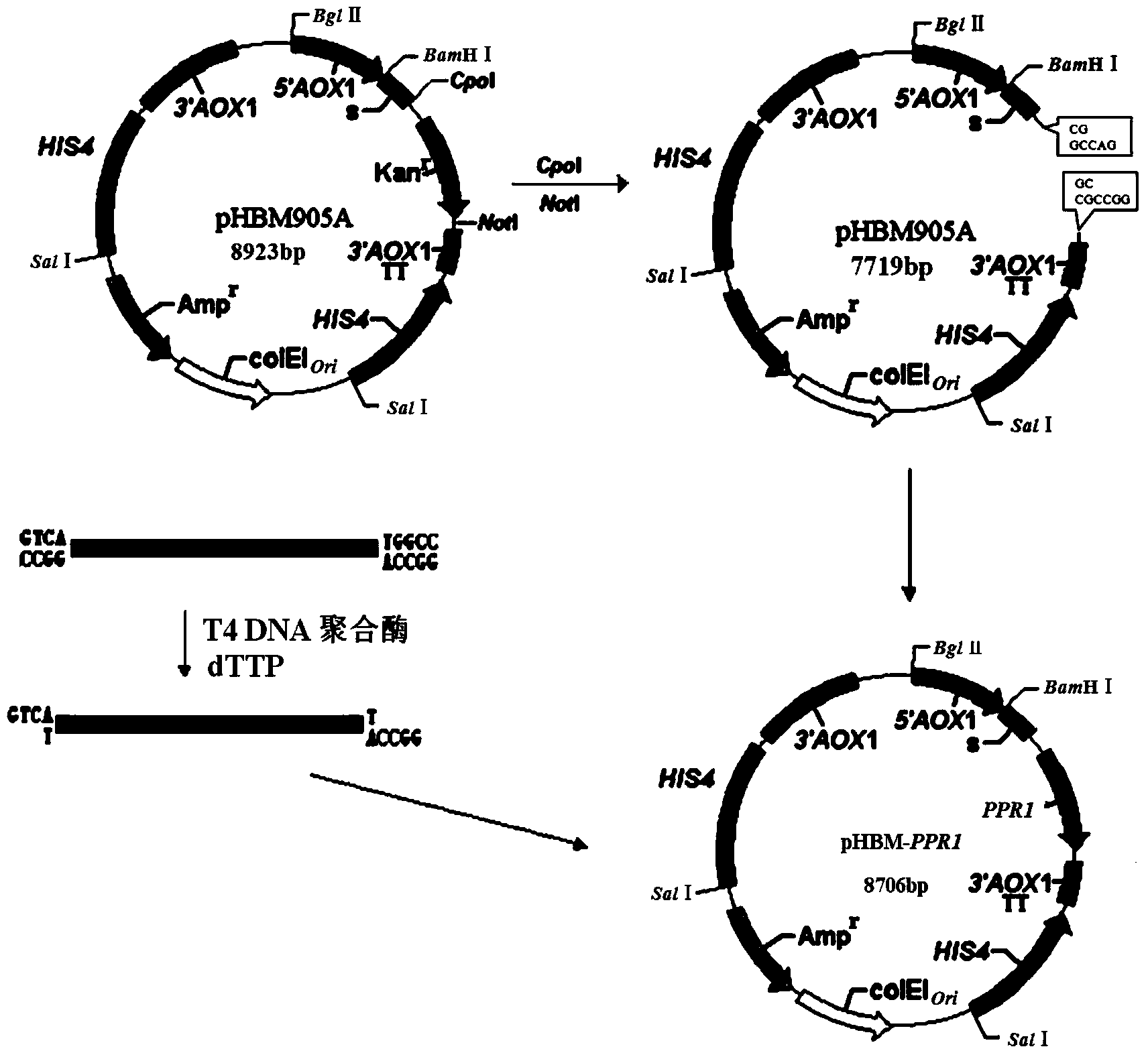
[0064] The Pichia pastoris expression vector pHBM905A (8923bp) was digested with Cop I and Not I, separated by agarose electrophoresis, and the large fragment (7719bp) was recovered by tapping the gel.
[0065] The PCR product (1005bp) synthesized in Example 2 was treated with T4 DNA polymerase in the presence of dTTP, and then ligated with the large fragment recovered from rubber tapping to obtain the Pichia pastoris recombinant expression plasmid pHBM-905A-Pi-pprI (8706bp), Abbreviated as pHBM-Pi-pprI, the construction flow chart is shown in figure 2 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com