Construction method of saccharomyces cerevisiae strain with high yield of isobutanol
A technology of Saccharomyces cerevisiae and Saccharomyces cerevisiae, which is applied in the field of DNA recombination technology to prepare microorganisms and can solve problems such as imbalance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
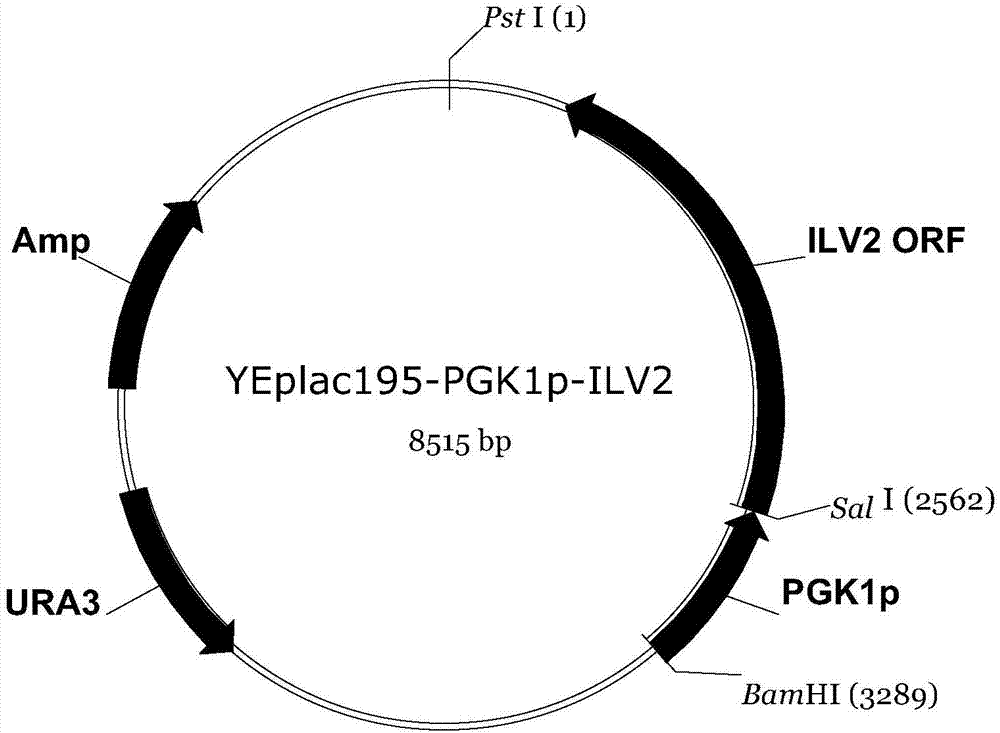
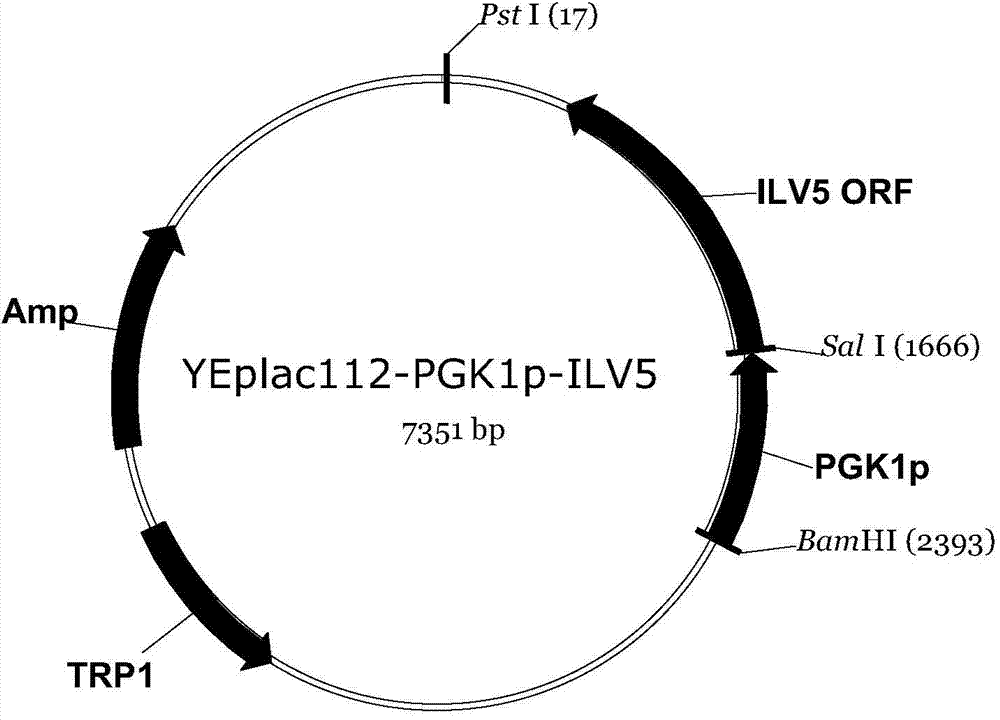
[0061] The host strain was Saccharomyces cerevisiae strain W303-1A (MATa leu2-3, 112ura3-1trp1-92his3-11, 15ade2-1can1-100 (Thomas and Rothstein, 1989)), and at the same time, it was transferred into the overexpressing ILV2, ILV3, ILV5, ZWF1 gene Plasmids YEplac195-PGK1p-ILV2, YEplac181-PGK1p-ILV3 and YEplac112-PGK1p-ZWF1-PGK1p-ILV5,
[0062] The first step, construction of plasmids for overexpressing Saccharomyces cerevisiae ILV2, ILV3, ILV5 and ZWF1 genes
[0063] (1.1) Preparation of Saccharomyces cerevisiae W303-1A chromosome: Inoculate Saccharomyces cerevisiae W303-1A into 5 mL of YPD medium, vibrate at 30°C overnight to obtain a culture solution, divide the culture solution into two 1.5 mL In the centrifuge tube, centrifuge at 12000rpm for 30s, take the supernatant, add 0.5mL water to resuspend, repeatedly suck the tip of the pipette to disperse the sedimentation of bacteria, then centrifuge at 12000rpm for 30s, collect the bacteria, and put them in Add 200 μL of bacter...
Embodiment 2
[0084] Except that the host bacterium is Saccharomyces cerevisiae strain and is selected as an industrial Saccharomyces cerevisiae strain, the others are the same as in Example 1.
[0085] The raw materials used in the above examples are all known and commercially available, and the operating techniques used are within the grasp of those skilled in the art.
Embodiment 3
[0087] Divide with T 4 Ligase to connect YEplac195-PGK1p and ILV2ORF genes: In a 1.5mL centrifuge tube, add 10×T 4 Ligase buffer 2 μL, T 4 The amount of ligase 0.2 μL, YEplac195-PGK1p and ILV2ORF gene fragments added was 4 μL, and then the total volume was made up to 20 μL with sterile water, and the ligation product was obtained at 16°C for 1.5 h; 4 Ligase to connect YEplac181-PGK1p and ILV3ORF genes: In a 1.5mL centrifuge tube, add 10×T 4 Ligase buffer 2 μL, T 4 Add 0.2 μL of ligase, 4 μL of YEplac181-PGK1p and ILV3ORF gene fragments, add sterile water to a total volume of 20 μL, and connect at 16°C for 1.5 h to obtain the ligation product; 4 Ligase to connect YEplac112-PGK1p and ILV5ORF genes: In a 1.5mL centrifuge tube, add 10×T 4 Ligase buffer 2 μL, T 4 Add 0.2 μL of ligase, 4 μL of YEplac112-PGK1p and ILV5ORF gene fragments, add sterile water to a total volume of 20 μL, and connect at 16°C for 1.5 h to obtain the ligation product; 4 Ligase the YIplac211-PGK1p and Z...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com