Novel fusion protein of mispairing binding protein and cellulose binding domain 3 and method thereof for removing errors in DNA (Desoxvribose Nucleic Acid) synthesis at high flux
A fusion protein and binding protein technology, applied in the field of high-throughput correction of DNA synthesis errors, can solve problems such as DNA synthesis errors, poor quality, complex oligonucleotide library components, etc., to achieve cost reduction, low price, high error With the effect of removing ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0040] Preparation of amorphous cellulose:
[0041] 0.2g of microcrystalline cellulose and 0.6mL of water were thoroughly mixed in a 50mL centrifuge tube to prepare a cellulose suspension, and then 10mL of ice-cold concentrated phosphoric acid (>85%) was added. Add slowly while mixing vigorously. This cellulose mixture will quickly become transparent. Place on ice for 1 h with occasional stirring. Then 40 mL of ice-cold water was added in four portions. Stir vigorously when adding to avoid lumps of cellulose. The precipitated cellulose was centrifuged at 5000 x g for 20 min at 4°C. Discard the supernatant, resuspend the pellet in ice water, centrifuge again, and discard the supernatant, repeat 4 times to remove phosphoric acid. Add 1mL2MNa 2 CO 3 Add 40mL of ice-cold water to the cellulose suspension to neutralize residual phosphoric acid, centrifuge to remove the supernatant, and wash with water twice. If the pH of the cellulose is 5-7, the preparation of amorphous cel...
Embodiment 1
[0051] The preparation of embodiment 1, eMutS and tMutS protein
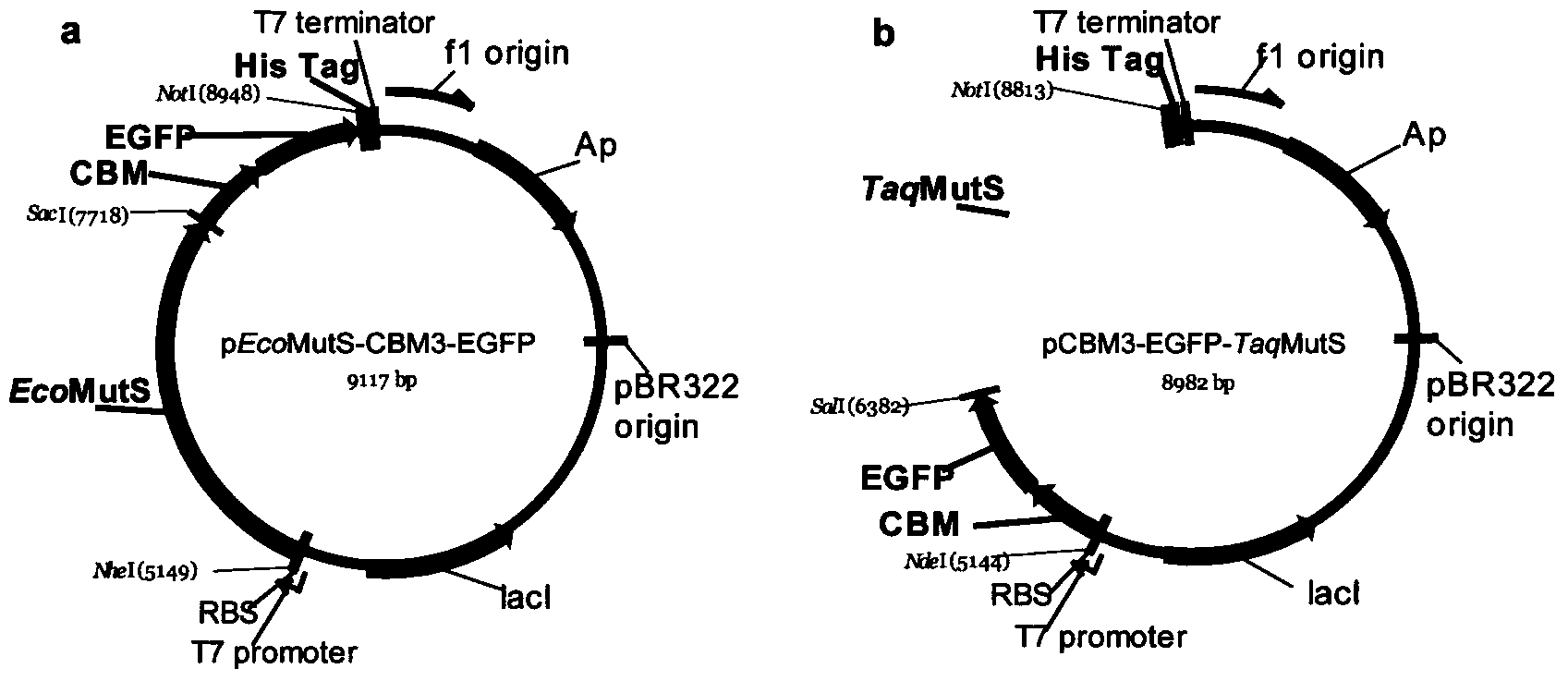
[0052] 1. Construction of eMutS and tMutS protein expression vectors
[0053] 1). Acquisition of EcoMutS gene:
[0054] Constructed with the plasmid pET-32a-eMutS (inserting the EcoMutS gene (GenBank: HG738867.1) into the EcoRI and HindIII sites of pET-32a, see Bi Lijun et al., AnthonyE.G.Cass (2002) DNA mismatch repair gene High-efficiency expression of mutS and identification of expression product activity. Acta Bioengineering 18(5):536-540) as a template, design specific amplification primers, and use PrimeSTAR HS DNA polymerase (Bao Biological Engineering (Dalian) Co., Ltd.) for PCR , the obtained product fragment is an EcoMutS-NS fragment containing the EcoMutS gene ("-NS" indicates that the fragment contains restriction sites N: NheI and S: SacI, the following are similar).
[0055] PCR system of EcoMutS-NS gene:
[0056]
[0057]
[0058] PCR program
[0059]
[0060] 2). Obtaining of TaqMutS...
Embodiment 2
[0140] Embodiment 2: the construction of MCSC:
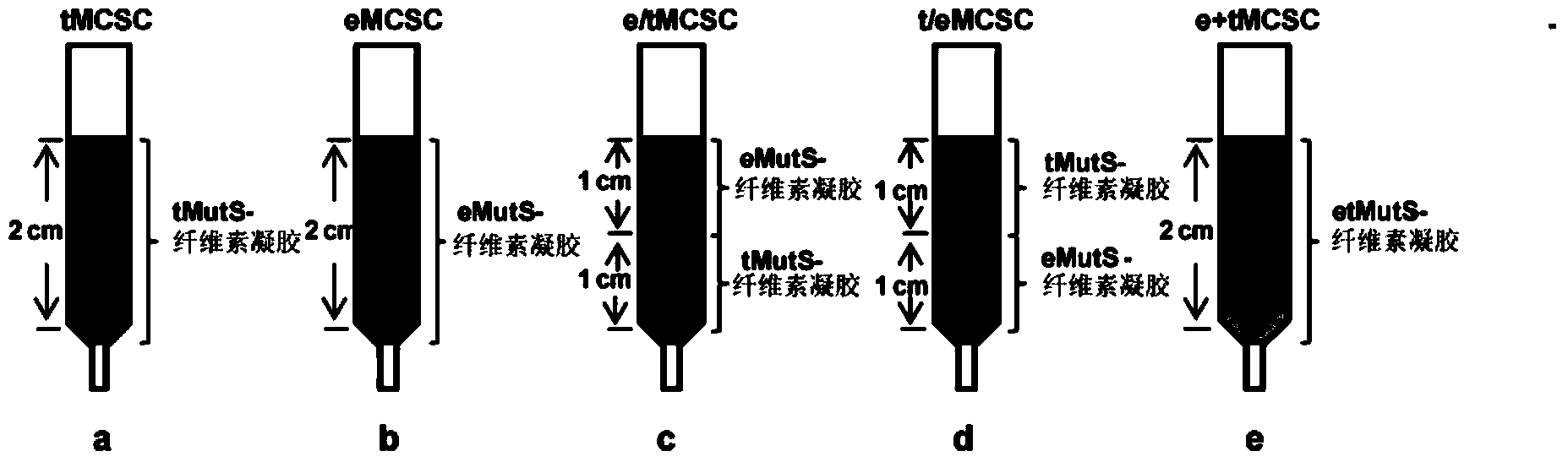
[0141] The MutS protein fused with CBM3 can be immobilized on cellulose by utilizing the stable and specific binding ability between CBM3 and amorphous cellulose, and then the cellulose immobilized with MutS protein can be packed into an empty column to form MutS cellulose column (MCSC).
[0142] The specific operation steps are:
[0143]In order to assemble a standard 2cm-high MCSC, 1ml of cellulose (20mg / ml) was mixed with 1200pmol of MutS fusion protein, combined at room temperature for 10 minutes, and the cellulose bound to MutS was loaded into an empty column (0.4cm×7cm, Sangon Bioengineering (Shanghai) Co., Ltd.) formed a column with a height of 2 cm and a diameter of 0.4 cm. After the column is stabilized, add 1 ml of binding buffer [5 mM MgCl2, 100 mM KCl, 20 mM Tris-HCl (pH7.6), 1 mM DTT] to wash away unbound MutS protein. According to the above-mentioned MCSC packing method, a total of five MCSCs ( figure 1 ), the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com