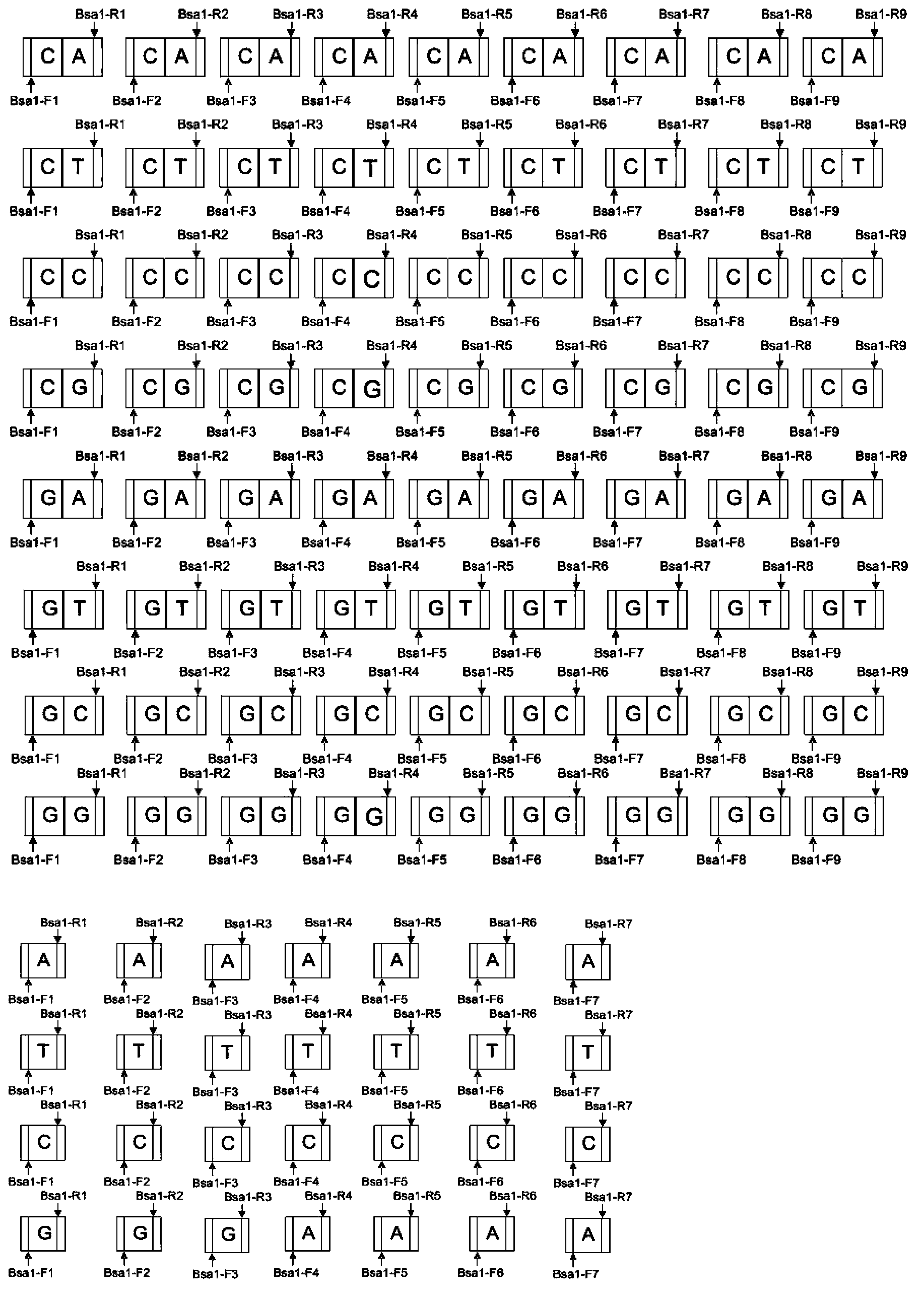DNA (Deoxyribonucleic Acid) library and method for constructing transcription activator like effector nuclease plasmids
A DNA library and transcription activation technology, applied in the field of genetic engineering, can solve the problems of difficulty, cumbersome operation, and high cost, and achieve the effects of avoiding damage and degradation, simple operation steps, and improved production technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Connection between TALENs recognition modules and construction of recombinant vector
[0042] 1. Acquisition of 20 identification modules (modular)
[0043] (1) The synthesis recognizes 4 single bases A, T, C, G and 16 double bases AA, AT, AC, AG, TA, TT, TC, TG, CA, CT, CC, CG, GA, 20 identification modules for GT, GC, GG NI, NG, HD, NN, NI-NI, NI-NG, NI-HD, NI-NN, NG-NI, NG-NG, NG-HD, NG-NN, The sequences of HD-NI, HD-NG, HD-HD, HD-NN, NN-NI, NN-NG, NN-HD, and NN-NN are shown in Table 1, as shown in SEQ ID NO: 1-20, respectively.
[0044] Table 1
[0045]
[0046]
[0047]
[0048] 2. Make 172 plasmid libraries by adding restriction sites and linkers to the basic modules
[0049] (1) PCR amplification to add enzyme-cut recognition sequences and connection adapters
[0050] F1, R1; F2, R2; F3, R3; F4, R4; F5, R5; F6, R6; F7, R7; F8, R8; F9, R9 as primers, T vector containing 16 double modules as template Do PCR, a total of 16×9=144;
[0051] Us...
Embodiment 2
[0102] connect identifiable bases
[0103] Fragment 1: TALEN of CGCGCGCGCGCGCGCGCGCGT (SEQ ID NO: 47), Fragment 2 CCCACTCCCCATCCAGT (SEQ ID NO: 48).
[0104] (1) First select the required recognition module from the PCR library:
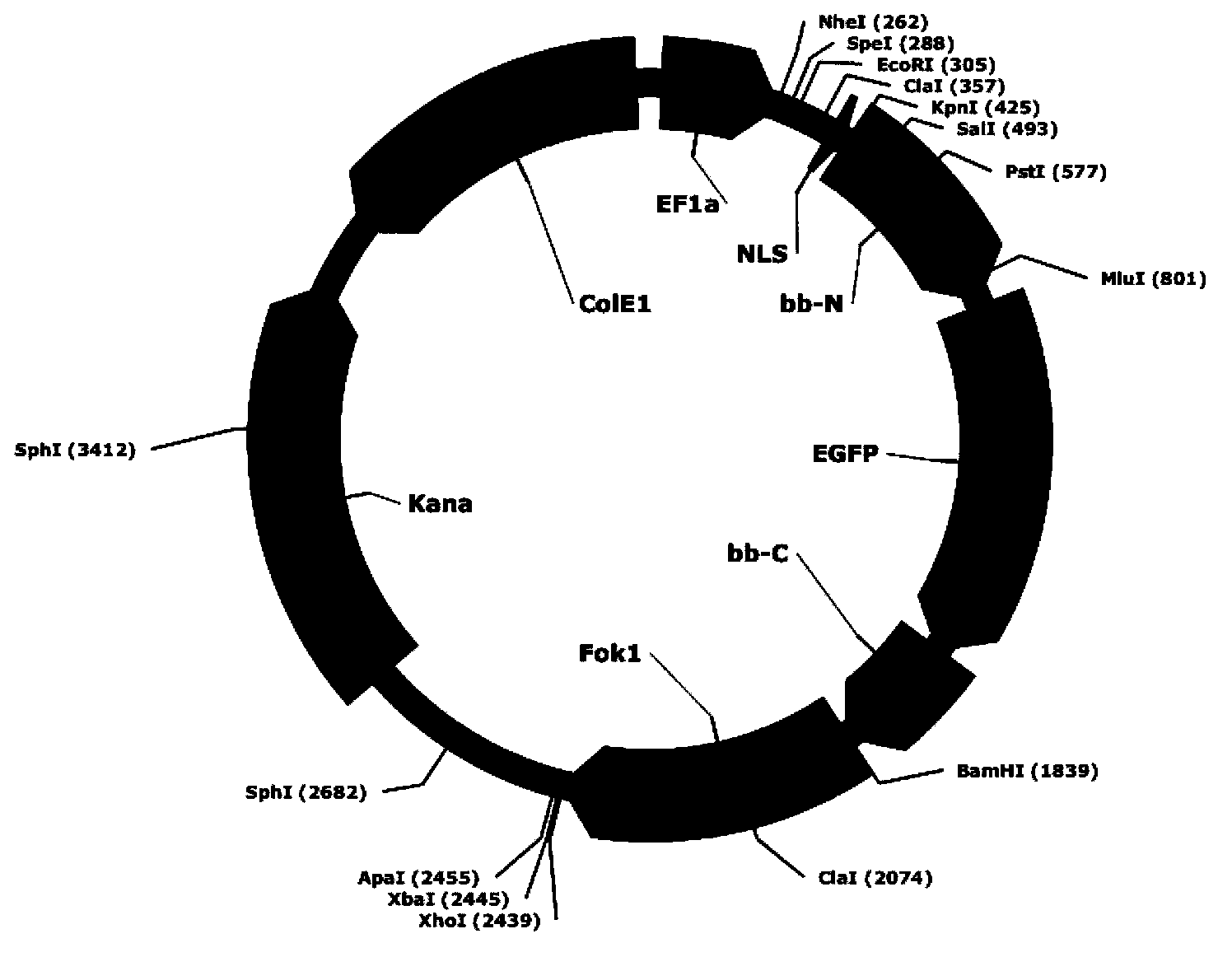
[0105] CGCGCGCGCGCGCGCGCGT selects modules CG-1, CG-2, CG-3, CG-4, CG-5, CG-6, CG-7, CG-8, CG-9; this connection vector is pEF1a-NLS-TALE backbone- Fok1(R)-pA
[0106] CACTCCCCATCCAGT select modules C-1, A-2, C-3, TC-4, CC-5, CA-6, TC-7, CA-8, GT-9. The connection vector is pEF1a-NLS-TALE backbone-Fok1(L)-IRES-PURO-pA
[0107] (2) According to the following connection system:
[0108] Vector: 150ng
[0109] Modulars: 50ng / modular
[0110] Bsa I (NEB): 1ul
[0111] T4Ligase (fermentas): 1ul
[0112] T4Buffer (NEB): 2ul
[0113] h 2 O: make up 20ul
[0114] The ligation program was: 5 min at 37°C; 15 cycles at 16°C for 10 min; 10 min at 80°C.
[0115] (3) Then plasma-safenuclease digestion: 1h.
[0116] (4) Transform Trans-T1 competent, pi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com