Site-directed modification method for DNA viral genome
A DNA virus and fixed-point transformation technology, which is applied in plant gene improvement, recombinant DNA technology, genetic engineering, etc., can solve the problems of low recombination rate, complicated operation of inserting foreign fragments, and difficult point-directed mutation of DNA virus genome, so as to achieve improved transformation The effect of improving efficiency and mutation rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
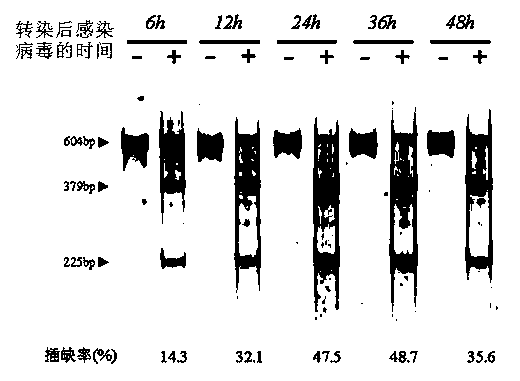
Image
Examples
Embodiment 1
[0044] Example 1: Targeted Knockout of Herpes Simplex Virus Genome TK Gene
[0045] Use the strain of herpes simplex virus type I (HSV1), which can be isolated and preserved by existing methods, for example, infect Vero cells with the fluid from a patient with clinical herpes simplex type I, and amplify to obtain HSV1 8F virus , wherein, 8F is the code name of the virus strain in the preparation process.
[0046] The site-directed knockout transformation method of the herpes simplex virus genome TK gene is realized through the following steps:
[0047] Step 1: After digesting and purifying the AG230 plasmid with BbsI, use T4 DNA ligase (NEB) to connect into the DNA via P109 (5'- CAC CGA GGG CGC AAC GCC GTA CGT -3') and P110 (5'- AAA CAC GTA CGG CGT TGC GCC CTC -3') The double-stranded insert sequence formed by the annealing of two synthetic primers was used to construct plasmid pCW206, and the plasmid pCW206 was extracted by an endotoxin-free plasmid extraction kit for use; ...
Embodiment 2
[0056] Example 2: Rapid site-specific insertion of exogenous red fluorescent protein into recombinant adenovirus vector
[0057]The recombinant adenovirus ADV-152 expressing green fluorescent protein was used. This strain used stratagene’s AdEasy? After infecting AD293 cells, ADV-152 recombinant adenovirus can be prepared and amplified. In this example, green fluorescent protein is replaced with red fluorescent protein. Among them, 152 in ADV-152 is the code name of the virus strain in the preparation process.
[0058] The transformation method of point-directed insertion of exogenous red fluorescent protein of recombinant adenovirus vector is realized through the following steps:
[0059] Step 1: After digesting and purifying the AG230 plasmid with BbsI, use T4 DNA ligase (NEB) to connect to the DNA via P069 (5'- CAC CGC TGA AGC ACT GCA CGC CGT -3') and P070 (5'- AAA CAC GGC GTG CAG TGC TTC AGC -3') The double-stranded insert sequence formed by the annealing of two synthet...
Embodiment 3
[0066] Example 3: Large fragment deletion of specific sequence of recombinant adenovirus
[0067] The recombinant adenovirus ADV-152 expressing green fluorescent protein was used. This strain used stratagene’s AdEasy? After infecting AD293 cells, the virus ADV-152 can be prepared and amplified.
[0068] The large-segment deletion transformation method of the specific sequence of the recombinant adenovirus genome is realized through the following steps:
[0069] Step 1: After digesting and purifying the AG230 plasmid with BbsI, use T4 DNA ligase (NEB) to connect into the DNA via P067 (5'- CAC CGG AGC GCA CCA TCT TCT TCA -3') and P068 (5'- AAA CTG AAG AAG ATG GTG CGC TCC -3') The double-stranded insert sequence formed by the annealing of two synthetic primers was used to construct the plasmid pCW174; then the AG230 plasmid was digested and purified with BbsI, and then ligated into the pCW174 via T4 DNA ligase (NEB) (5'- CAC CGC TGA AGC ACT GCA CGC CGT -3') and P070 (5'- AAA CA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com