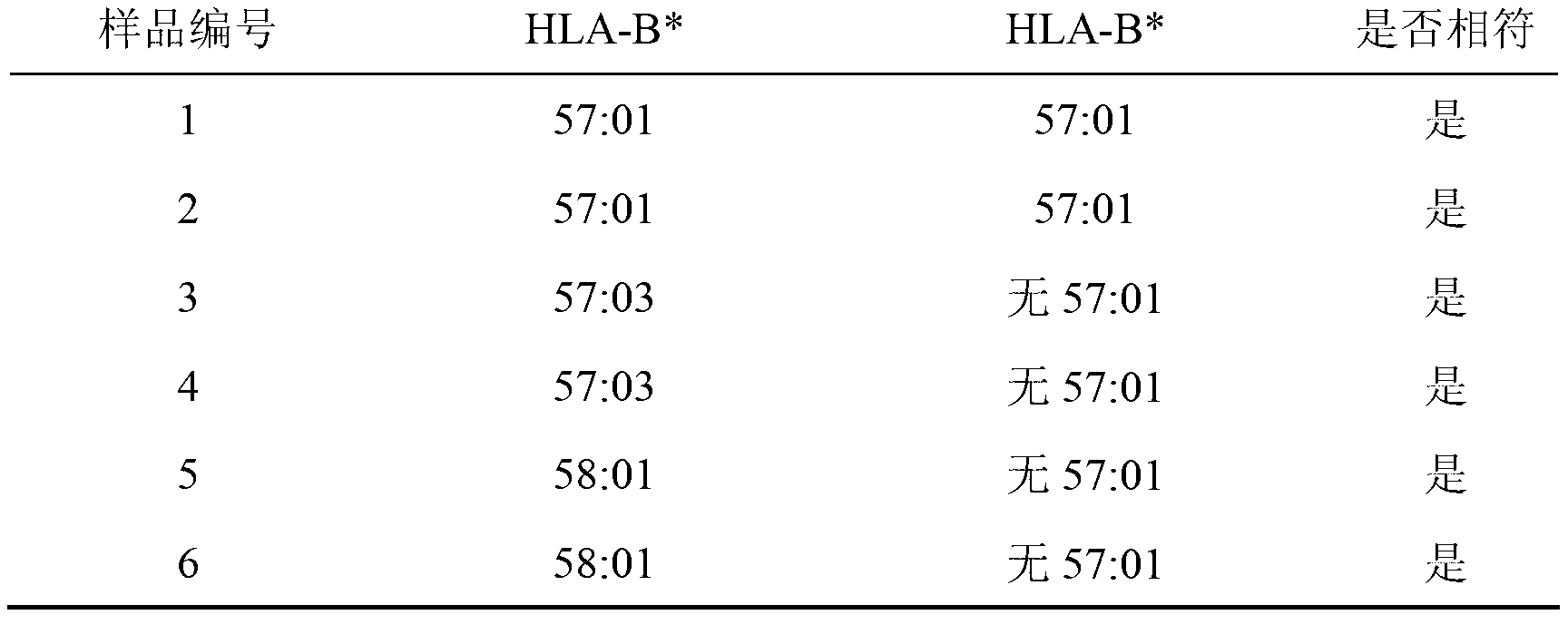Assay kit for detecting human leukocyte antigen-B (HLA-B)*57:01 and HLA complex P5 (HCP5) alleles
A technology of HLA-B and alleles, applied in the biological field, can solve the problems of high detection cost and long time, achieve the effect of low experimental cost, prevent false negative or false positive, and save workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] The design of the PCR primers of the kit of the present invention In this embodiment, all the HLA allele sequence data required for PCR primer design are taken from the international HLA database (IMGT / HLA), the database website: http: / / www .ebi.ac.uk / ipd / imgt / hla / . Sequence alignment was performed using the software Sequence Alignment Tool of the database. The primers were designed manually, and the designed primers were tested by BLAST in the GenBank database of the NIH in the United States, and it was confirmed that the primers could specifically bind to the HLA-B*57:01 allele. In this kit, the key is that the primers need to specifically amplify the HLA-B*57:01 gene in a specific PCR buffer system environment, that is, the primers have "sequence-specific SSP". There are two solutions in this primer design, one is to actually test primers of different lengths to specifically bind the target gene; the other is to introduce a mutant base at the 3' end of the primer to...
Embodiment 2
[0055] Embodiment 2: the preparation of kit of the present invention
[0056] 1. Synthesis of primers: The primers were synthesized by Shanghai Yingjun Biotechnology Co., Ltd. The sequences of the primers are SEQ ID NO: 1-7, and the specific sequences are as described in Example 1.
[0057] 2. Preparation of PCR reaction mixture: Mix primers 1-7, dNTPs, dye cresyl red, and Taq enzyme buffer. The concentration of the detection primer (SEQ ID NO:1-5) was 0.5 μM, the concentration of the internal reference primer (SEQ ID NO:6-7) was 0.2 μM; the concentration of dNTP was 0.2 mM; the concentration of cresol red was 0.01% ( wt / vol); PCR buffer system: Tris-HCl 10mM, potassium chloride 50mM, magnesium chloride 1.5mM, gelatin 0.001% (wt / vol).
[0058] The above-mentioned PCR reaction mixture and Taq enzyme are subpackaged and packaged to form a kit.
Embodiment 3
[0059] Example 3: Detection of HLA-B*57:01 and HCP5rs2395029SNP in samples
[0060] Take a PCR tube, add 8 μl of PCR reaction mixture to each tube, and then add 1 μl of 50ng of genomic DNA extracted from 6 copies of HLA standard cell lines (all HLA standard cell lines were purchased from "IHWG Cell and DNA Bank") , and then add 0.30-0.5uTaq enzyme. After adding the sample, mix the reaction mixture evenly, centrifuge briefly, and carry out the PCR reaction.
[0061] The reaction conditions of PCR are: 95°C for 5min; followed by 30 cycles according to the following procedure, 95°C for 30s, 60°C for 30s, 72°C for 90s; finally 72°C for 5min, cooling to 4°C for gel electrophoresis detection.
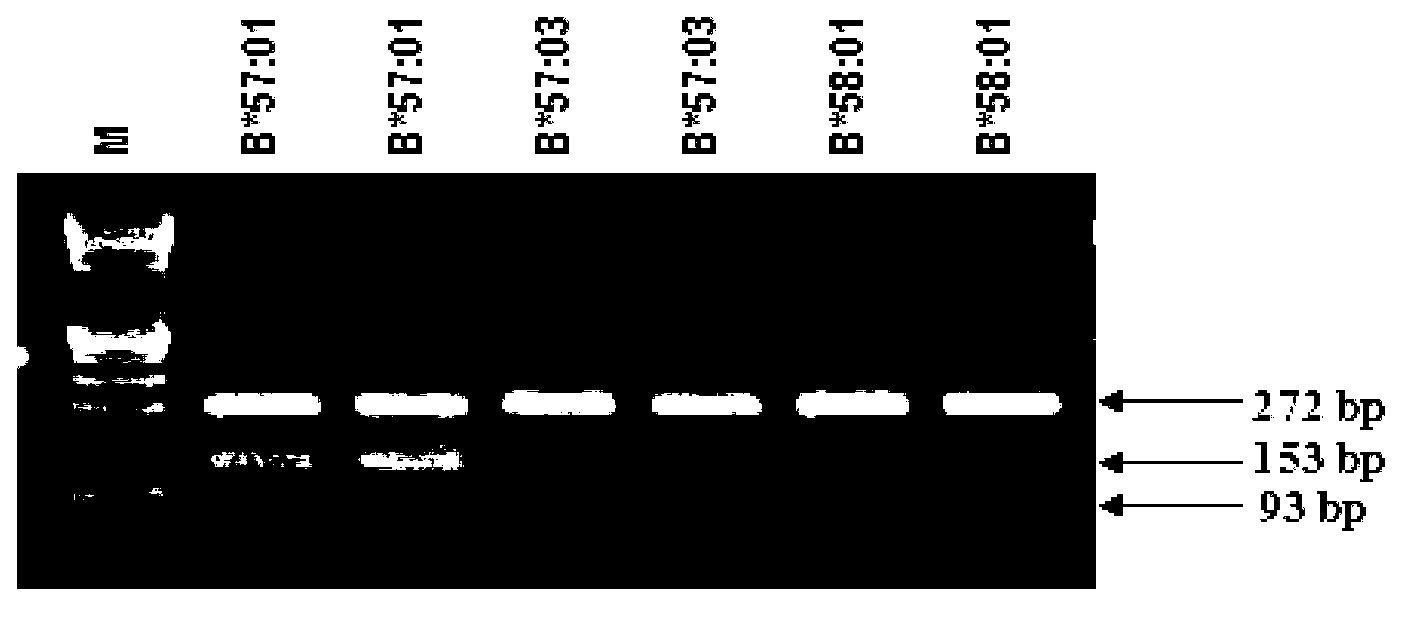
[0062] Using 0.5×TBE buffer, prepare 3% agarose gel. Take 5 μl of the PCR product and spot it directly into the well of the gel. Use 0.5×TBE buffer solution, electrophoresis at 10V / cm for 15-20 minutes, and then take pictures and record under ultraviolet light (see figure 1 ).
[0063] J...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com