Molecular marker related to micropterus salmoides hatchability and application thereof
A technology for largemouth bass and hatchability, which is applied in the field of animal genetic engineering, can solve the problems affecting the screening effect of parents with high hatchability of largemouth bass, and achieve the effects of improving relative hatchability, rapid detection, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Screening of insertion / deletion sites
[0031] 1. Experimental fish
[0032] In November 2008, 170 largemouth bass samples were randomly collected from the breeding base of Jiujiang Town, Nanhai District, Foshan City, Guangdong Province for the screening of gene mutation sites and the correlation analysis of growth traits. The body weight, body length and body height of each largemouth bass were measured. At the same time, blood was collected from the tail vein. The ratio of anticoagulant (ACD) to blood volume was 6:1, and DNA was extracted with Blood&Cell Culture DNA Kit (Clontech).
[0033] 2. Amplification of the GHRH promoter sequence
[0034] The cDNA sequence of the growth hormone releasing hormone (GHRH) gene of largemouth bass (GenBank accession number: HQ640678) was compared with the complete sequence of the known fish GHRH gene, and primers P1 and P2 were designed (Table 1), Promoter for amplification of GHRH. According to the operation of t...
Embodiment 2
[0054] A method for assisting in screening high-hatchability largemouth bass parents, comprising the steps of:
[0055] 1. Design of primers
[0056] According to the flanking sequence of a 66 bp recessive lethal deletion mutation site on the promoter sequence of the growth hormone releasing hormone (GHRH) gene of largemouth bass, primers were designed and synthesized as follows (see figure 1 ):
[0057] P1: GGCCCGGGCTGGTCTGTTAAATACA (SEQ ID NO: 3)
[0058] P2: GCGCAACACAAGAGCACATTGCTT (SEQ ID NO: 4)
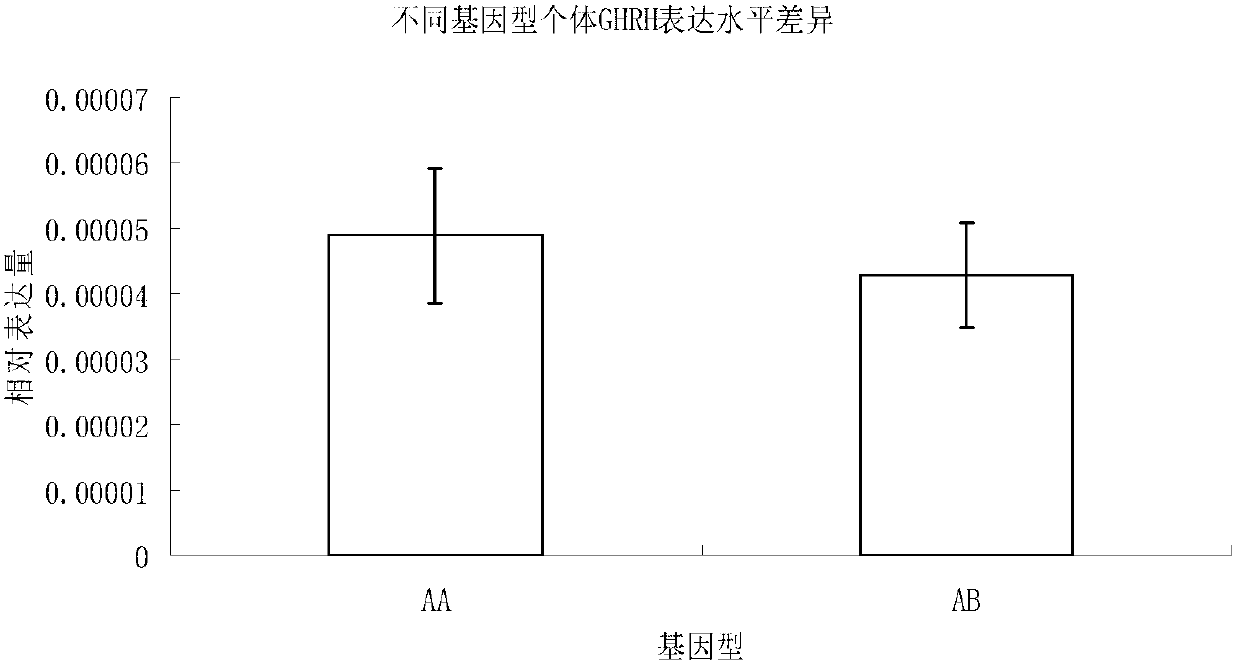
[0059] Individuals with AA genotype are expected to amplify 1 DNA band with a size of 316bp; individuals with AB genotype are expected to amplify 2 DNA bands with a size of 316bp and 250bp.
[0060] 2. Template DNA extraction and PCR amplification
[0061] (1) Take 3 mg of fin ray tissue of the broodstock to be tested and cut it into pieces, add 0.5 mL of lysate (10 mmol / L Tris-HCl; 0.1 mol / L EDTA; 0.5% SDS; 30 mg / L RNase; 100 mg / L protease K, pH8.0), digested at 55°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com