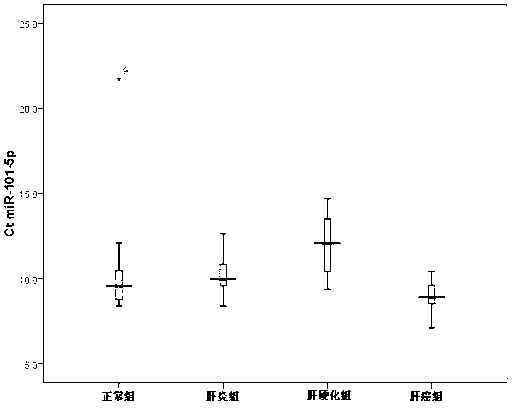Method and composition based on tiny RNA for liver cirrhosis and early liver cancer diagnosis
A diagnostic composition, tiny technology, applied in biochemical equipment and methods, microbial assay/examination, etc., capable of solving problems such as sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1 research object
[0045] A total of 122 subjects were used in this study, which were divided into patient group and healthy normal people group, and the patient group was further divided into three groups: chronic hepatitis B group (hereinafter referred to as hepatitis B group, 30 cases), hepatitis B-related cirrhosis group ( Hereinafter referred to as liver cirrhosis group, 30 cases) and hepatitis B related liver cancer group (hereinafter referred to as liver cancer group, 32 cases); healthy normal people group (hereinafter referred to as normal group, 30 cases). The study complies with the requirements of the Declaration of Helsinki, and volunteers have been informed about the use of their blood, tissue samples and clinical data.
[0046] All patients in the patient group were HBsAg positive and had no other types of liver diseases such as chronic hepatitis C, alcoholic liver disease, autoimmune liver disease or metabolic liver disease. All data were o...
Embodiment 2
[0047] Example 2 Serum Sample Acquisition and Sample RNA Extraction
[0048] For the patient group, 10ml of peripheral blood was collected from the patients before liver biopsy or operation and stored in serum test tubes. For the normal group, 10ml of peripheral blood from healthy normal people were collected and stored in serum test tubes. The tubes were centrifuged at 1500 g for 10 min at 4°C. The serum was then aliquoted and centrifuged at 3000 g for 15 min at 4°C to remove cell debris and other residual cells. Serum samples were stored at -80°C.
[0049]All RNA was extracted from 200 μl of serum using the Serum Total RNA Extraction Kit (QuantoBio, Beijing, CN). First, add 200 μl lysis buffer (Lysis Buffer) to each sample, vortex evenly; then add 40 μl lysis enhancer solution (Lysis Enhancer Solution), shake and mix by hand for 15 seconds; then add 440 μl acidified phenol: chloroform (Acidfied Phenol :Chloroform), shake and mix by hand for 30s. Centrifuge at 16,000 g...
Embodiment 3
[0050] Embodiment 3 Reverse transcription obtains cDNA
[0051] cDNA is reverse transcribed from RNA by polyadenylation. Mix 10pg-1μg of total RNA template with 2μl 10× buffer (buffer), 2μl dATP (10mM), 0.5μl polyA polymerase (NEB), 0.5μl ribonuclease (RNase) inhibitor (Promega) and no ribonuclease Water (RNase freewater) (Promega) was mixed to a final volume of 20 μl, and incubated at 37°C for 1h. Then add 1 μl 0.5 μg / μl RT primer (QuantoBio, Beijing, CN) to the reaction tube, incubate at 70°C for 5 minutes and immediately incubate on ice for at least 2 minutes to break the secondary structure of RNA and primers. Finally, the above 20 μl reaction mixture was mixed with 4 μl 5× buffer (buffer), 1 μl dNTP (10 mM) (Sigma), 0.5 μl M-MLV reverse transcriptase (Promega), 0.5 μl ribonuclease (RNase) inhibitor (Promega) , 10 μl polyA reaction mixture and 4 μl RNase free water (Promega), incubated at 42°C for 1h. Undiluted cDNA templates were stored at -20°C for future use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com