Method used for detecting serum miRNA of patients with cancer based on short nucleotide chain connection
A short nucleotide and chain connection technology, which is applied in biochemical equipment and methods, microbiological determination/inspection, etc., to achieve the effect of simple operation, accurate and reliable detection results, and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
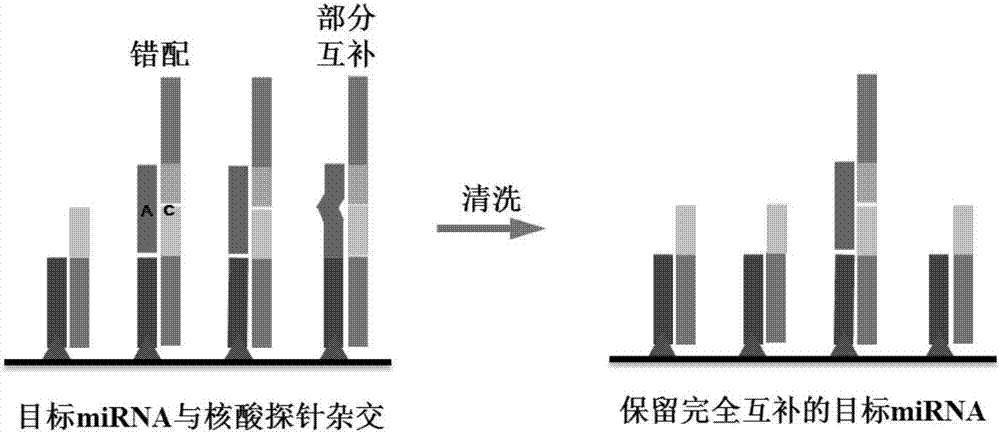
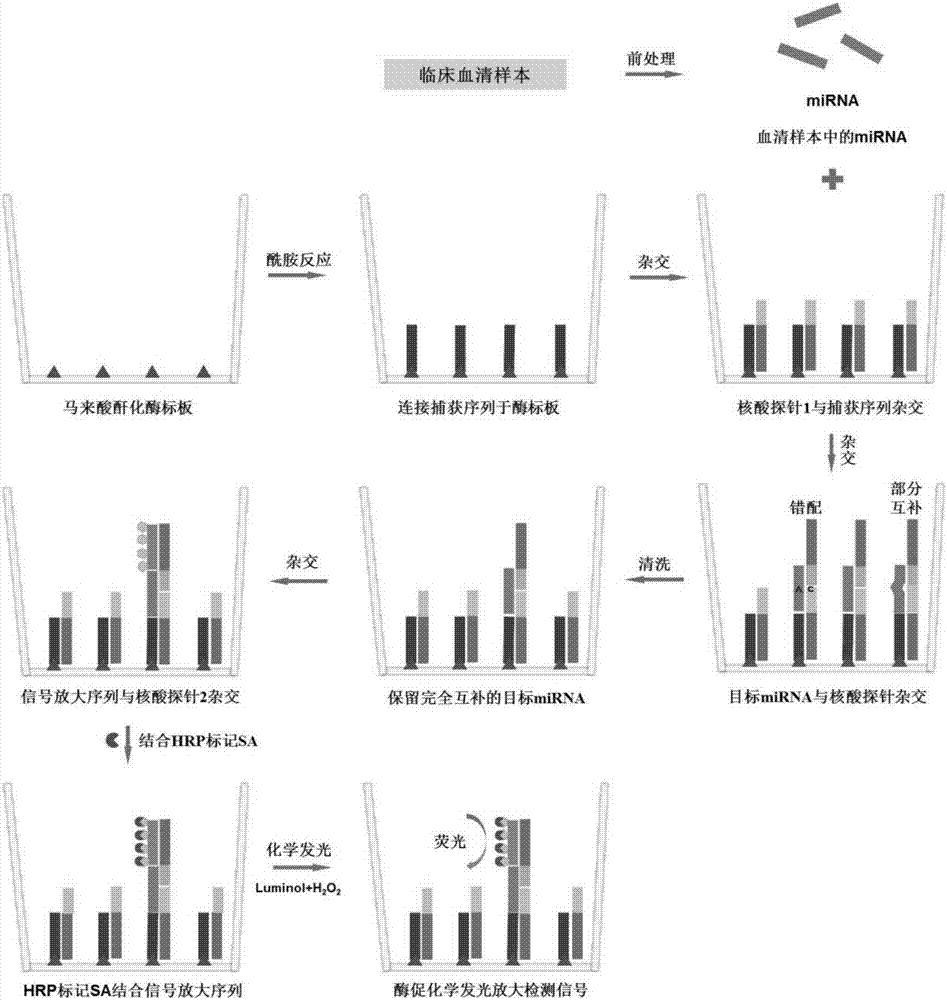
[0032]A method for detecting serum miRNA in cancer patients based on short nucleotide chain ligation, the establishment and optimization of high-throughput SOL-based technology for detecting serum miRNA: pretreatment of serum samples, and capture sequence, nucleic acid probe 1, nucleic acid Probe 2, signal amplification sequence hybridization, and then use SA-HRP to catalyze the chemiluminescent substrate solution to generate strong fluorescence, and determine the expression level of target miRNA in serum by detecting the fluorescence intensity; systematically optimize serum processing time, nucleic acid hybridization temperature, signal Amplify the number of Biotin on the sequence and other conditions, and establish a new method for miRNA detection with strong specificity, accurate and reliable results, and easy operation. miRNA; at the same time, using this miRNA detection method to detect miR-16 and miR-21 in the serum samples of 20 cases of healthy controls and 34 cases of ...
specific Embodiment 2
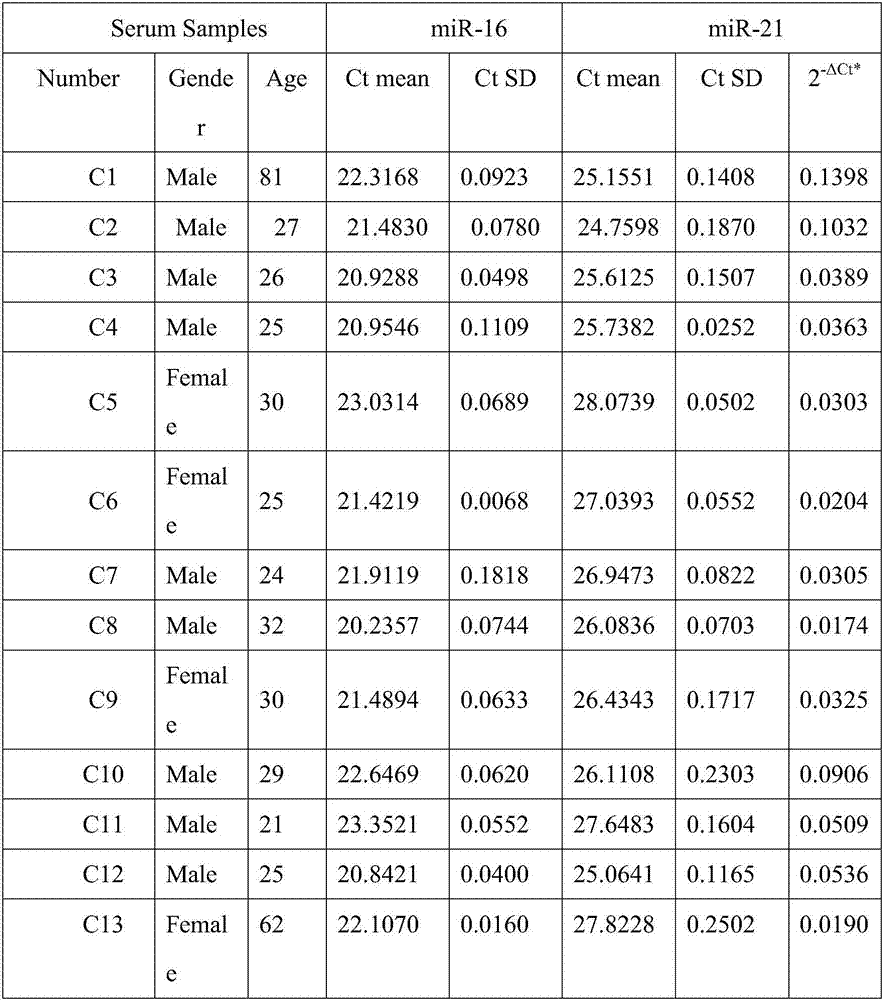
[0033] A method based on short nucleotide chain ligation to detect serum miRNA in cancer patients, using SOL technology to detect specific miRNA markers in serum for early diagnosis of lung cancer: using the new method of SOL detection combined with well-explored serum pretreatment methods, Two miRNAs, miR-16 and miR-21, were detected in the serum samples of 20 healthy controls and 34 NSCLC patients with different stages, ages and genders; miR-16 with relatively stable expression was used as an internal reference substance, and miR-21, which was specifically highly expressed in the serum of NSCLC patients, was used as a miRNA marker. ) detected miR-16 and miR-21 in the collected serum samples by fluorescent quantitative PCR, and the Ct value detected by miR-21 was normalized with the Ct value of internal reference miR-16, and miR-21 was compared with internal reference miR Multiples of -16 are represented by 2-ΔCt, see Table 1-2.
[0034] Table 1 Real-time quantitative PCR de...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com