A mutant of cutinase and its preparation method
A cutinase and mutant technology, applied in the field of genetic engineering and enzyme engineering, can solve the problems of poor thermal stability, unsuitable application and low hydrolysis efficiency of fungal cutinase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Method for determining mutation sites of cutinase mutants of Thermobifida fusca.
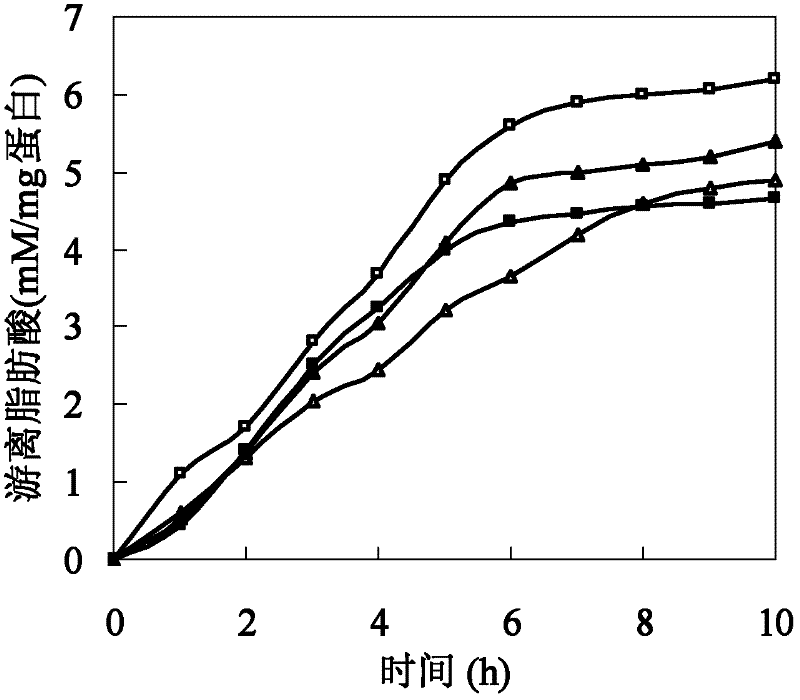
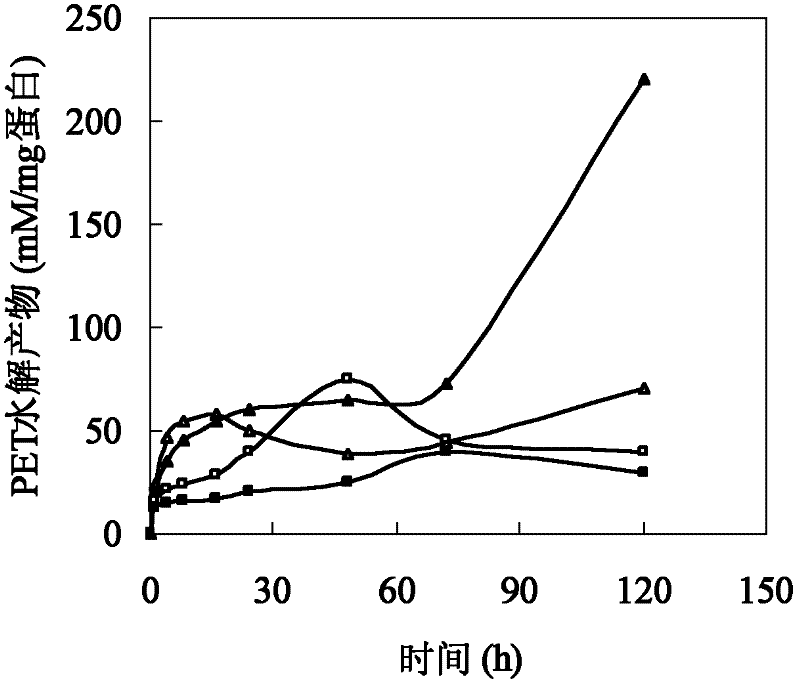
[0026] Polyesters such as cutin are hydrophobic macromolecules composed of a large number of fatty acids. The size of amino acids around the active center of cutinase affects the combination of polyester macromolecules and active centers. In addition, the hydrophobicity of amino acids affects the substrate binding site and substrate. Therefore, based on the cutinase mimic structure, proceeding from the above two aspects, determine the mutation site and then perform site-directed mutation on the cutinase gene. Based on the analysis of the simulated structure of the cutinase of Thermobifida fusca, three sets of mutation series were designed: (1) The amino acid isoleucine I218 located directly above the active center of serine occupies a large space, which has a large impact on the active center and The combination of macromolecular substrates forms a large steric hindrance, so it...
Embodiment 2
[0027] Example 2: The preparation method of the cutinase mutant of Thermobifida fusca.
[0028] I218A-pet20b(+), W195 / F249-pet20b(+), Q132A / T101A-pet20b(+) mutant plasmids were constructed by rapid PCR site-directed mutagenesis.
[0029] Construction of I218A-pet20b(+), W195-pet20b(+), Q132A-pet20b(+) mutant plasmids using the CUT-pet20b(+) plasmid as a template (Chen Jian, Wu Jing, Chen Sheng. A heat-resistant cutinase And its coding gene and expression. Application No. 200710026074.2), use the mutant primers of I218A, W195A and Q132A to obtain the product with a size of about 4500bp by PCR respectively, transform the Escherichia coli JM109 competent cells after the PCR product is treated with Dpn I, and select the transformed Subsequence verification.
[0030] The W195 / F249-pet20b(+), Q132A / T101A-pet20b(+) mutant plasmids were constructed using the verified correct W195-pet20b(+), Q132A-pet20b(+) plasmids as templates, and F249A and T101A as mutation primers , PCR amplifie...
Embodiment 3
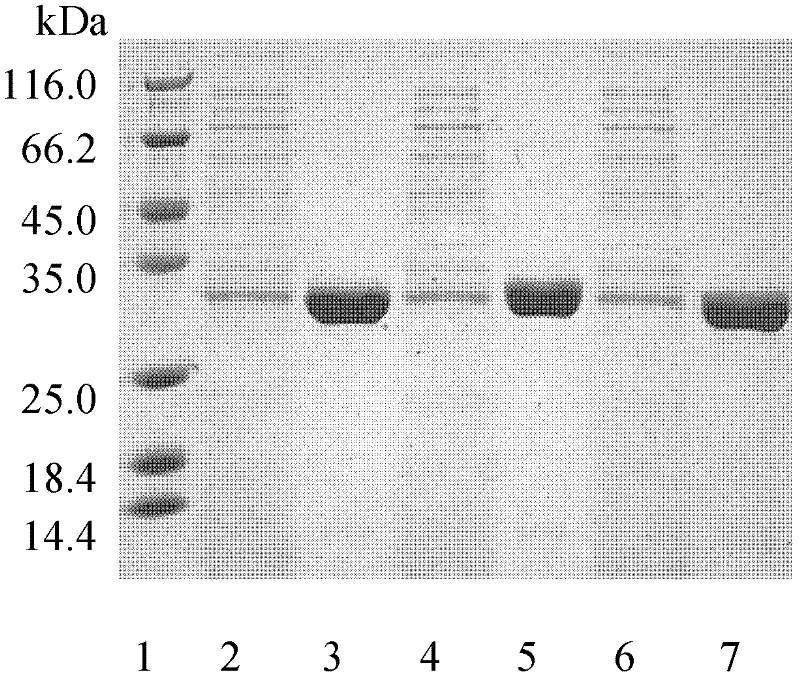
[0053] Example 3: Method for expression and purification of cutinase mutants from Thermobifida fusca.
[0054] The mutant plasmids I218A-pet20b(+), W195 / F249-pet20b(+), Q132A / T101A-pet20b(+) were transformed into E. coli BL21(DE3) cells, and the transformants were selected for sequencing verification. The positive transformant after verification was in TB medium (glycerol 5g / L, peptone 12g / L, yeast extract 24g / L, K 2 HPO 4 12.54g / L, KH 2 PO 4 2.31g / L) in 37°C liquid culture overnight, then inserted into TB fermentation liquid medium and cultured at 37°C for 3 hours, then induced with 4 mg / L IPTG (isopropylthioβD-galactoside), cooled to 25°C and incubated at constant temperature for 80 hours .
[0055] The fermentation broth was centrifuged at 10,000 rpm for 20 minutes at 4°C to remove bacteria. The supernatant was collected through an activated carbon column. Add 70% solid ammonium sulfate to the clear solution passing through the activated carbon column for salting ou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com