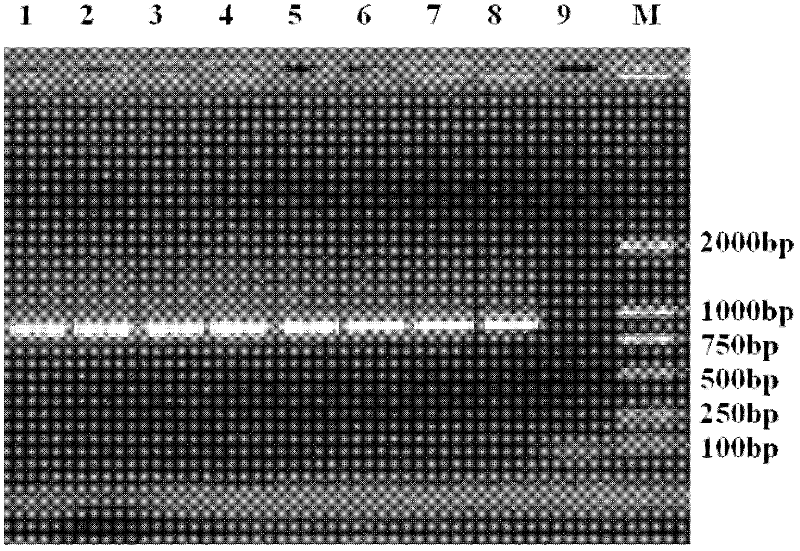
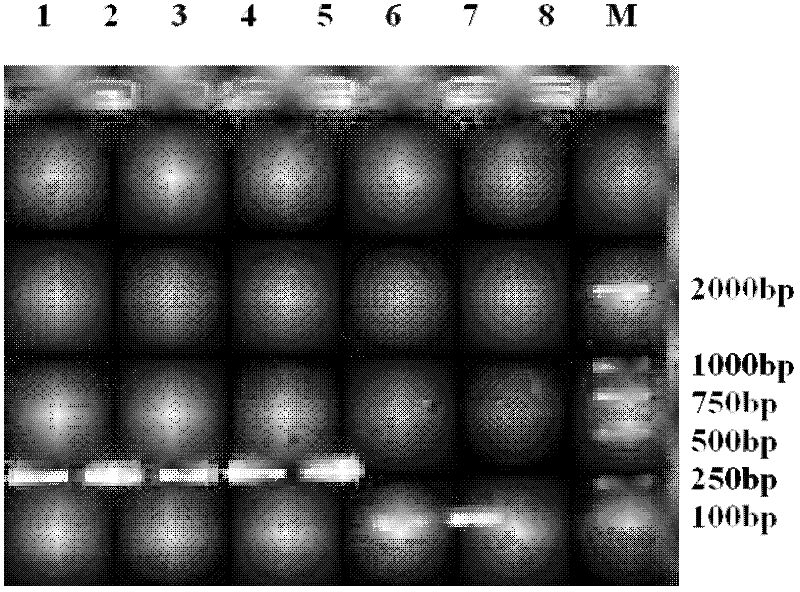
Method for detecting TilletiafuscaEII&EV and T.bromi(Brockm.)Brockm by using double polymerase chain reaction (PCR) primers
A smut and double technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of time-consuming and laborious germination experiments, unfavorable rapid detection of ports, etc., to save reagents and time, rapid detection, highly specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1: the germination of experimental material teliospore and the cultivation of mycelia
[0042] The materials involved in the experiment include Tilletia A total of 7 strains (mainly hyphae and teliospores) of 2 similar species of the genus were obtained from the Phytosanitary Laboratory of the Animal, Plant and Food Testing Center of the Tianjin Entry-Exit Inspection and Quarantine Bureau. Zao1, Zao2, ZaoL, Poa5, and Poa6 were imported from the United States. LMC307 and LMC312 were exchanged specimens, and all strains were cultured from teliospores. The relevant information of the tested materials is shown in Table 1.
[0043]
[0044] Table 1 Test materials
[0045] serial number the code Latin scientific name the host years source 1 LMC307 Tilletia fusca Vulpia microstachys 1995 USA 2 LMC312 T. fusca V. octoflova 1995 USA 3 Zao1 Tilletia bromi Poa pratensis 2003 USA 4 Zao2 T....
Embodiment 2
[0047] Embodiment 2: the extraction of hyphal genome DNA
[0048] Mycelial DNA was extracted using Shanghai Sangon Genomic DNA Purification Kit (No. SK1252). The extracted genomic DNA was dissolved in 70 μL 1×TE, and the remaining hyphae were stored at -70°C for use.
Embodiment 3
[0049] Embodiment 3: the picking and breaking of teliospores
[0050] Place a 1mm square cover glass on the glass slide, drop about 0.5 μL of 10×PCR buffer on it, puncture the gall with a dissecting needle, pick about 3-10 teliospores and put them in 10×PCR buffer, cover with an approximate size Gently rub the coverslip with tweezers, confirm the rupture of spores under the microscope, put the superimposed two coverslips together into the bottom of the PCR tube containing 4.5 μL 10×PCR buffer, cover the tube cap, and No teliospores were added and only PCR Buffer was used as a negative control.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com